Abstract
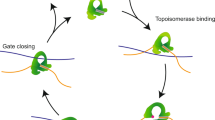
Cells must remove all entanglements between their replicated chromosomal DNAs to segregate them during cell division. Entanglement removal is done by ATP-driven enzymes that pass DNA strands through one another, called type II topoisomerases. In vitro, some type II topoisomerases can reduce entanglements much more than expected, given the assumption that they pass DNA segments through one another in a random way1. These type II topoisomerases (of less than 10 nm in diameter) thus use ATP hydrolysis to sense and remove entanglements spread along flexible DNA strands of up to 3,000 nm long. Here we propose a mechanism for this, based on the higher rate of collisions along entangled DNA strands, relative to collision rates on disentangled DNA strands. We show theoretically that if a type II topoisomerase requires an initial ‘activating’ collision before a second strand-passing collision, the probability of entanglement may be reduced to experimentally observed levels. This proposed two-collision reaction is similar to ‘kinetic proofreading’ models of molecular recognition2,3.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rybenkov,V. V., Ullsperger,C., Vologodskii,A. V. & Cozzarelli,N. R. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science 277, 690–693 (1997).
Hopfield,J. J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl Acad. Sci. USA 71, 4135–4139 (1974).
Ninio,J. Kinetic amplification of enzyme discrimination. Biochimie 57, 587–595 (1975).
Frank-Kamenetskii,M. D., Lukashin,A. V. & Vologodskii,A. V. Statistical mechanics and topology of polymer chains. Nature 258, 398–402 (1975).
Klenin,K. V., Vologodskii,A. V., Anshelevich,V. V., Dykhne,A. M. & Frank-Kamenetskii,M. D. Effect of excluded volume on topological properties of circular DNA. J. Biomol. Struct. Dyn. 5, 1173–1185 (1988).
Shaw,S. Y. & Wang,J. C. Knotting of a DNA chain during ring closure. Science 260, 533–536 (1993).
Rybenkov,A. V., Cozzarelli,N. R. & Vologodskii,A. V. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl Acad. Sci. USA 90, 5307–5311 (1993).
Vologodskii,A. V. in RECOMB 98: Proceedings of the second annual international conference on computational molecular biology 266–269 (Association for Computing Machinery, New York, 1998).
Deguchi,T. & Tsurusaki,K. A statistical study of random knotting using the Vassiliev invariants. J. Knot Theory Ramific. 3, 321–353 (1994).
Roca,J. & Wang,J. C. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell 77, 609–616 (1994).
Roca,J. & Wang,J. C. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71, 833–840 (1992).
Roca,J., Berger,J. M. & Wang,J. C. On the simultaneous binding of eukaryotic DNA topoisomerase II to a pair of double-stranded DNA helices. J. Biol. Chem. 268, 14250–14255 (1993).
Harkins,T. T. & Lindsley,J. E. Pre-steady-state analysis of ATP hydrolysis by Saccaromyces cerevisiae DNA topoisomerase II. 1. A DNA-dependent burst in ATP hydrolysis. Biochemistry 37, 7292–7299 (1998).
Harkins,T. T. & Lindsley,J. E. Pre-steady-state analysis of ATP hydrolysis by Saccaromyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry 37, 7299–7312 (1998).
Worcel,A. & Burgi,E. On the structure of the folded chromosome of Escherichia coli. J. Mol. Biol. 71, 127–147 (1972).
Sinden,R. R. & Pettijohn,D. E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl Acad. Sci. USA 78, 224–228 (1981).
Acknowledgements
We thank D. Chatenay, N. R. Cozzarelli, G. B. Mindlin, V. Rybenkov, E. D. Siggia, A. V. Vologodskii and E. L. Zechiedrich for discussions. J.F.M. and J.Y. acknowledge the support of the NSF, the Research Corporation, the Petroleum Research Fund, and the Whitaker Foundation. M.O.M. acknowledges support of the Sloan Foundation and the Mathers Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yan, J., Magnasco, M. & Marko, J. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature 401, 932–935 (1999). https://doi.org/10.1038/44872
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/44872
This article is cited by
-
Ratcheting synthesis
Nature Reviews Chemistry (2023)
-
Biological Implications of Dynamical Phases in Non-equilibrium Networks
Journal of Statistical Physics (2016)
-
Controlling gene expression by DNA mechanics: emerging insights and challenges
Biophysical Reviews (2016)
-
The dynamic interplay between DNA topoisomerases and DNA topology
Biophysical Reviews (2016)
-
Controlling gene expression by DNA mechanics: emerging insights and challenges
Biophysical Reviews (2016)