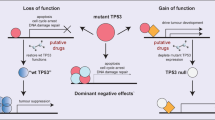
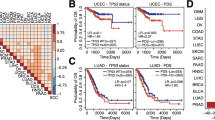
Abstract
TP53 is probably the most extensively studied tumour-suppressor gene, and patients with TP53 mutations are known to have a poor outcome. However, inconsistencies in the analysis of TP53 status, and failure to realize that different mutations behave in different ways, prevent us from effectively applying our vast knowledge of this protein in clinical practice. What simple steps can be taken to ensure that patients benefit from our understanding of TP53?
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vousden, K. H. p53. Death star. Cell 103, 691–694 (2000).
Vogelstein, B., Lane, D. & Levine, A. J. Surfing the p53 network. Nature 408, 307–310 (2000).
Soussi, T., Dehouche, K. & Béroud, C. p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum. Mutat. 15, 105–113 (2000).
Moll, U. M., Laquaglia, M., Benard, J. & Riou, G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc. Natl Acad. Sci. USA 92, 4407–4411 (1995).
Oliner, J. D., Kinzler, K. W., Meltzer, P. S., Georges, D. L. & Vogelstein, B. Amplification of a gene encoding a p53 associated protein in human sarcomas. Nature 358, 80–83 (1992).
Crook, T. et al. Clonal p53 mutation in primary cervical cancer- association with human-papillomavirus-negative tumours. Lancet 339, 1070–1073 (1992).
Soengas, M. S. et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature 409, 207–211 (2001).
Bell, D. W. et al. Heterozygous germ line hCHK2 mutations in Li–Fraumeni syndrome. Science 286, 2528–2531 (1999).
Rotman, G. & Shiloh, Y. ATM: a mediator of multiple responses to genotoxic stress. Oncogene 18, 6135–6144 (1999).
Baker, S. J. et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244, 217–221 (1989).
Takahashi, T. et al. p53- a frequent target for genetic abnormalities in lung cancer. Science 246, 491–494 (1989).
Nigro, J. M. et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 342, 705–708 (1989).
Dowell, S. P., Wilson, P. O. G., Derias, N. W., Lane, D. P. & Hall, P. A. Clinical utility of the immunocytochemical detection of p53 protein in cytological specimens. Cancer Res. 54, 2914–2918 (1994).
Varley, J. M. et al. Characterization of germline TP53 splicing mutations and their genetic and functional analysis. Oncogene 20, 2647–2654 (1999).
Gu, J., Kawai, H., Wiederschain, D. & Yuan, Z. M. Mechanism of functional inactivation of a Li–Fraumeni syndrome p53 that has a mutation outside of the DNA-binding domain. Cancer Res. 61, 1741–1746 (2001).
Hashimoto, T. et al. p53 null mutations undetected by immunohistochemical staining predict a poor outcome with early-stage non-small cell lung carcinomas. Cancer Res. 59, 5572–5577 (1999).
Skaug, V. et al. p53 mutations in defined structural and functional domains are related to poor clinical outcome in non-small cell lung cancer patients. Clin. Cancer Res. 6, 1031–1037 (2000).
Tomizawa, Y. et al. Correlation between the status of the p53 gene and survival in patients with stage I non-small cell lung carcinoma. Oncogene 18, 1007–1014 (1999).
Schiller, J. H. et al. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: a Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. J. Clin. Oncol. 19, 448–457 (2001).
Hartmann, A., Blaszyk, H., Kovach, J. S. & Sommer, S. S. The molecular epidemiology of p53 gene mutations in human breast cancer. Trends Genet. 13, 27–33 (1997).
Kern, S. E. et al. Mutant p53 proteins bind DNA abnormally in vitro. Oncogene 6, 131–136 (1991).
El-Deiry, W. S., Kern, S. E., Pientenpol, J. A., Kinzler, K. W. & Vogelstein, B. Definition of a consensus binding site for p53. Nature Genet. 1, 45–49 (1992).
Kern, S. E. et al. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256, 827–830 (1992).
Milner, J. & Medcalf, E. A. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 65, 765–774 (1991).
Dittmer, D. et al. Gain of function mutations in p53. Nature Genet. 4, 42–46 (1993).
Halevy, O., Michalovitz, D. & Oren, M. Different tumor-derived p53 mutants exhibit distinct biological activities. Science 250, 113–116 (1990).
Harvey, M. et al. A mutant p53 transgene accelerates tumour development in heterozygous but not nullizygous p53 deficient mice. Nature Genet. 9, 305–311 (1995).
Blandino, G., Levine, A. J. & Oren, M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 18, 477–485 (1999).
Rowan, S. et al. Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. EMBO J. 15, 827–838 (1996).
Forrester, K. et al. Effects of p53 mutants on wild-type p53-mediated transactivation are cell type dependent. Oncogene 10, 2103–2111 (1995).
Cho, Y. J., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. Crystal structure of a p53 tumor suppressor DNA complex: understanding tumorigenic mutations. Science 265, 346–355 (1994).
Ory, K., Legros, Y., Auguin, C. & Soussi, T. Analysis of the most representative tumour-derived p53 mutants reveals that changes in protein conformation are not correlated with loss of transactivation or inhibition of cell proliferation. EMBO J. 13, 3496–3504 (1994).
Hinds, P. W. et al. Mutant p53 DNA clones from human colon carcinomas cooperate with Ras in transforming primary rat cells: a comparison of the 'Hot Spot' mutant phenotypes. Cell Growth Differ. 1, 571–580 (1990).
Selivanova, G. et al. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nature Med. 3, 632–638 (1997).
Bullock, A. N., Henckel, J. & Fersht, A. R. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene 19, 1245–1256 (2000).
Wong, K. B. et al. Hot-spot mutants of p53 core domain evince characteristic local structural changes. Proc. Natl Acad. Sci. USA 96, 8438–8442 (1999).
Berns, E. et al. Mutations in residues of TP53 that directly contact DNA predict poor outcome in human primary breast cancer. Br. J. Cancer 77, 1130–1136 (1998).
Kucera, E. et al. Prognostic significance of mutations in the p53 gene, particularly in the zinc-binding domains, in lymph node- and steroid receptor positive breast cancer patients. Eur. J. Cancer 35, 398–405 (1999).
Borresen, A. L. et al. TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc-binding domains. Gene Chromosom. Cancer 14, 71–75 (1995).
Borresen Dale, A. L. et al. TP53 and long-term prognosis in colorectal cancer: mutations in the L3 zinc-binding domain predict poor survival. Clin. Cancer Res. 4, 203–210 (1998).
Goh, H. S., Yao, J. & Smith, D. R. p53 point mutation and survival in colorectal cancer patients. Cancer Res. 55, 5217–5221 (1995).
Liu, G. et al. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc. Natl Acad. Sci. USA 97, 4174–4179 (2000).
Chappuis, P. O. et al. Prognostic significance of p53 mutation in breast cancer: frequent detection of non-missense mutations by yeast functional assay. Int. J. Cancer 84, 587–593 (1999).
Smith, P. D. et al. Novel p53 mutants selected in BRCA-associated tumours which dissociate transformation suppression from other wild-type p53 functions. Oncogene 18, 2451–2459 (1999).
de Cremoux, P. et al. p53 mutation as a genetic trait of typical medullary breast carcinoma. J. Natl Cancer Inst. 91, 641–643 (1999).
Yang, A. & McKeon, F. p63 and p73: p53 mimics, menaces and more. Nature Rev. Mol. Cell Biol. 1, 199–207 (2000).
Ikawa, S., Nakagawara, A. & Ikawa, Y. p53 family genes: structural comparison, expression and mutation. Cell Death Differ. 6, 1154–1161 (1999).
Levrero, M. et al. Structure, function and regulation of p63 and p73. Cell Death Differ. 6, 1146–1153 (1999).
Hibi, K. et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl Acad. Sci. USA 97, 5462–5467 (2000).
Strano, S. et al. Physical and functional interaction between p53 mutants and different isoforms of p73. J. Biol. Chem. 275, 29503–29512 (2000).
Marin, M. C. et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nature Genet. 25, 47–54 (2000).
Gaiddon, C., Lokshin, M., Ahn, J., Zhang, T. & Prives, C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell Biol. 21, 1874–1887 (2001).
Agami, R., Blandino, G., Oren, M. & Shaul, Y. The tyrosine kinase c-ABL regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399, 806–809 (1999).
Ratovitski, E. A. et al. p53 associates with and targets ΔNp63 into a protein degradation pathway. Proc. Natl Acad. Sci. USA 98, 1817–1822 (2001).
Storey, A. et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature 393, 229–234 (1998).
Rosenthal, A. N. et al. p53 codon 72 polymorphism and risk of cervical cancer in UK. Lancet 352, 871–872 (1998).
Storey, A. et al. p53 polymorphism and risk of cervical cancer: reply. Nature 396, 532 (1998).
Lanham, S., Campbell, I., Watt, P. & Gornall, R. p53 polymorphism and risk of cervical cancer. Lancet 352, 1631–1631 (1998).
Helland, A. et al. p53 polymorphism and risk of cervical cancer. Nature 396, 530–531 (1998).
Zehbe, I. et al. p53 codon 72 polymorphism and various human papillomavirus 16 E6 genotypes are risk factors for cervical cancer development. Cancer Res. 61, 608–611 (2001).
Beckman, G. et al. Is p53 polymorphism maintained by natural selection? Hum. Hered. 44, 266–270 (1994).
Stolzenberg, M. C. et al. Germ-line exclusion of a single p53 allele by premature termination of translation in a Li–Fraumeni syndrome family. Oncogene 9, 2799–2804 (1994).
Ahrendt, S. A. et al. Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc. Natl Acad. Sci. USA 96, 7382–7387 (1999).
Bullock, A. N. & Fersht, A. R. Rescuing the function of mutant p53. Nature Reviews Cancer 1, 68–76 (2001).
Zhao, R. et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14, 981–993 (2000).
Yu, J. et al. Identification and classification of p53-regulated genes. Proc. Natl Acad. Sci. USA 96, 14517–14522 (1999).
Kostic, C. & Shaw, P. H. Isolation and characterization of sixteen novel p53 response genes. Oncogene 19, 3978–3987 (2000).
Kannan, K. et al. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20, 2225–2234 (2001).
Tokino, T. & Nakamura, Y. The role of p53-target genes in human cancer. Crit. Rev. Oncol. Hematol. 33, 1–6 (2000).
Hansen, R. & Oren, M. p53: from inductive signal to cellular effect. Curr. Opin. Genet. Dev. 7, 46–51 (1997).
May, P. & May, E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 18, 7621–7636 (1999).
Aas, T. et al. Specific p53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nature Med. 2, 811–814 (1996).
Geisler, S. et al. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res. 61, 2505–2512 (2001).
Takahashi, M. et al. Distinct prognostic values of p53 mutations and loss of estrogen receptor and their cumulative effect in primary breast cancers. Int. J. Cancer 89, 92–99 (2000).
Powell, B., Soong, R., Iacopetta, B., Seshadri, R. & Smith, D. R. Prognostic significance of mutations to different structural and functional regions of the p53 gene in breast cancer. Clin. Cancer Res. 6, 443–451 (2000).
Gentile, M., Jungestrom, M. B., Olsen, K. E., Soderkvist, P. & Wingren, S. p53 and survival in early onset breast cancer: analysis of gene mutations, loss of heterozygosity and protein accumulation. Eur. J. Cancer 35, 1202–1207 (1999).
Berns, E. M. et al. Complete sequencing of TP53 predicts poor response to systemic therapy of advanced breast cancer. Cancer Res. 60, 2155–2162 (2000).
Clausen, O. P. F. et al. Association of p53 accumulation with TP53 mutations, loss of heterozygosity at 17p13, and DNA ploidy status in 273 colorectal carcinomas. Diagn. Mol. Pathol. 7, 215–223 (1998).
Huang, C. et al. Mutations in exon 7 and 8 of p53 as poor prognostic factors in patients with non-small cell lung cancer. Oncogene 16, 2469–2477 (1998).
Vega, F. J. et al. p53 exon 5 mutations as a prognostic indicator of shortened survival in non-small-cell lung cancer. Br. J. Cancer 76, 44–51 (1997).
Erber, R. et al. TP53 DNA contact mutations are selectively associated with allelic loss and have a strong clinical impact in head and neck cancer. Oncogene 16, 1671–1679 (1998).
Kihara, C. et al. Mutations in zinc-binding domains of p53 as a prognostic marker of esophageal–cancer patients. Jpn J. Cancer Res. 91, 190–198 (2000).
Acknowledgements
We are grateful to D. Barnes, N. Basset-Seguin, E. M. J. J. Berns, A. L. Borresen, D. Brash, R. Camplejohn, R. Iggo, U. Moll, D. Sidransky and B. Vogelstein for critical reading of this manuscript. T.S. is grateful to B. Asselain and P. Viehl for helpful discussions. Our work is supported by grants from Association de Recherche contre le Cancer, Institut Curie, Ligue contre le Cancer (Comité de Paris) and Fondation de France.
Author information
Authors and Affiliations
Corresponding author
Related links
Rights and permissions
About this article
Cite this article
Soussi, T., Béroud, C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer 1, 233–239 (2001). https://doi.org/10.1038/35106009
Issue Date:
DOI: https://doi.org/10.1038/35106009