Key Points
-
This review discusses recent analyses of the genes required for autophagy ? intracellular bulk protein degradation ? in yeast, where two ubiquitin-like systems have now been revealed.
-
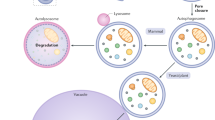
Light microscopy studies show that starved yeast cells take up their own cytoplasm into vacuoles through autophagic bodies. Closer analysis using electron microscopy show that these bodies form a double-membraned structure called the autophagosome, which subsequently fuses with the vacuole/lysosome. The whole process is topologically the same in mammals.
-
Screens in yeast defective in autophagy morphologically and biochemically revealed two sets of genes, the APG and AUT genes, respectively. A specific vacuolar enzyme biosynthetic pathway requires the cytosol-to-vacuole-targeting (CVT) genes. There is extensive overlap between the CVT genes and the APG/AUT genes.All apg mutants have defects at or before formation of the autophagosome.
-
Two ubiquitin-like systems have been discovered. The first ? the Apg12 conjugation system ? is unique in that Apg12 is synthesized as a mature form; it seems to have just one target, Apg5, and at steady state almost all Apg12 molecules are conjugated with Apg5.
-
The second ? Apg8/Aut7 ? is processed at its carboxy-terminal region by Aut2/Apg4. Apg8 exists in two forms, one of which is membrane bound through a phospholipid. This lipidation is mediated by ubiqutin-like system; Apg8 is activated by Apg7 and transferred to Apg3 and finally forms a conjugate with phosphatidylethaolamine (PE). Apg4 cleaves Apg8?PE, releasing Apg8 from membrane.
-
Morphological studies show that Apg8 localizes on the membrane of intermediate structures of the autophagosome; this transient association seems to be essential for formation of the autophagosome.
-
Both Apg12 and Apg8 are highly conserved, with apparent homologues in the worm, mammals and plants. In higher eukaryotes, Apg8 consists of a multigene family.
Abstract
Recent analyses of the genes required for autophagy ? intracellular bulk protein degradation ? in yeast have revealed two ubiquitin-like systems, both of which are involved in the membrane dynamics of the process. Molecular dissection of these systems is now revealing some surprises.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Notes
*Macroautophagy is generally a non-selective sequestration.But in some physiological situations, excess or unnecessary organelles areselectively degraded in a lysosome/vacuole. The best-studied case is peroxisomedegradation, called pexophagy by Daniel Klionsky. In the yeast Pichia pastoris, peroxisomes are sequestered through either macroautophagy or microautophagy,depending on the nutrient conditions40.
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425?479 (1998).
Klionsky, D. J. & Ohsumi, Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 1?32 ( 1999).
Baba, M., Takeshige, K., Baba, N. & Ohsumi, Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903?913 (1994).
Takeshige, K., Baba, M., Tsuboi, S., Noda, T. & Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301?311 (1992). This is the first report that yeast induces autophagy that is quite similar to that in mammals under various starvation conditions.
Kim, J. & Klionsky, D. J. Autophagy, the cytoplasm-to-vacuole-targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 69, 303?342 ( 2000).
Klionsky, D. J. & Emr, S. Autophagy as a regulated pathway of cellular degradation. Science 290, 1717?1721 (2000).
Baba, M., Osumi, M. & Ohsumi, Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 20, 465?471 (1995).
Tsukada, M. & Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169?174 (1993).
Thumm, M. et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae . FEBS Lett. 349, 275? 280 (1994).
Mizushima, N., Noda, T. & Ohsumi, Y. Apg16p is required for the function of the Apg12p?Apg5p conjugate in the yeast autophagy pathway. EMBO J. 18 , 3888?3896 (1999).
Kamada, Y. et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507? 1513 (2000).
Harding, T. M., Morano, K. A., Scott, S. V. & Klionsky, D. J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591?602 (1995).
Noda, T. et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465?480 ( 2000).
George, M. D. et al. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol. Biol. Cell 11, 969?982 (2000).
Yuan, W., Stromhaug, P. E. & Dunn, W. A. Jr Glucose-induced autophagy of peroxisomes in Pichia pastori requires a unique E1-like protein. Mol. Biol. Cell 10, 1353?1366 (1999).
Kim, J., Dalton, V. M., Eggerton, K. P., Scott, S. V. & Klionsky, D. J. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell 10, 1337? 1351 (1999).
Hutchins, M. U., Veenhuis, M. & Klionsky, D. J. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112, 4079?4087 (1999).
Muller, O. et al. Autophagic tubes. Vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 151, 519?528 (2000).
Suriapranata, I. et al. The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J. Cell Sci. 113, 4025? 4033 (2000).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395?398 ( 1998).This paper presented the first ubiquitin-like protein conjugation reaction, the Apg12 system, as being essential for autophagy.
Tanida, I. et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell 10, 1367? 1379 (1999).
Shintani, T. et al. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18, 5234? 5241 (1999).
Kirisako, T. et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435? 446 (1999).
Huang, W. P., Scott, S. V., Kim, J. & Klionsky, D. J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275, 5845?5851 (2000).
Lang, T. et al. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 17, 3597?3607 ( 1998).
Kirisako, T. et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263? 276 (2000).This showed that Apg8 associates with the intermediate membranes of the autophagosome by serial modification reactions.
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 489?493 ( 2000).This paper described the Apg8 lipidation system, in which the processed form of Apg8 is activated by a ubiquitin-like system and finally forms a conjugate with the membrane phospholipid, phosphatidylethanolamine. PubMed
Mizushima, N., Sugita, H., Yoshimori, T. & Ohsumi, Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 273, 33889?33892 (1998).
Mizushima, N. et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J.Cell Biol. (in the press).
Mann, S. S. & Hammarback, J. A. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol. Chem. 269, 11492?11497 (1994).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19 , 5720?5728 (2000). Two UBL systems, Apg12 and Apg8, are conserved in higher eukaryotes and this is the first report of the functional homologue of Apg8 in mammals.
Meyers, G., Stoll, D. & Gunn, M. Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J. Virol. 72, 4139?4148 (1998).
Sagiv, Y., Legesse-Miller, A., Porat, A. & Elazar, Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 19, 1494? 1504 (2000).
Legesse-Miller, A., Sagiv, Y., Glozman, R. & Elazar, Z. Aut7p, a soluble autophagic factor, participates in multiple membrane trafficking processes . J. Biol. Chem. 275, 32966? 32973 (2000).
Chen, L., Wang, H., Vicini, S. & Olsen, R. W. The γ-aminobutyric acid type A (GABAA) receptor-associated protein (GABARAP) promotes GABAA receptor clustering and modulates the channel kinetics. Proc. Natl Acad. Sci. USA 97, 11557? 11562 (2000).
Wang, H., Bedford, F. K., Brandon, N. J., Moss, S. J. & Olsen, R. W. GABAA-receptor-associated protein links GABAA receptors and the cytoskeleton. Nature 397, 69?72 ( 1999).
Paz, Y., Elazar, Z. & Fass, D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 275, 25445?25450 ( 2000).
Suzuki, K., Kirisako, T. & Ohsumi, Y. Localization of Apg5p and Apg8p/Aut7p reveals functional classes of APG genes and their interactions in autophagy. (submitted).
Wendland, B., Emr, S. D. & Riezman, H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr. Opin. Cell Biol. 10 , 513?522 (1998).
Tuttle, D. L. & Dunn, W. A. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 108, 25?35 (1995).
Huang, P. H. & Chiang, H. L. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J. Cell Biol. 136, 803?810 ( 1997).
Cuervo, A. M. & Dice, J. F. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275, 31505? 31513 (2000).
Scott, S. V., Baba, M., Ohsumi, Y. & Klionsky, D. J. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 138, 37?44 (1997).
Baba, M., Osumi, M., Scott, S. V., Klionsky, D. J. & Ohsumi, Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139 , 1687?1695 (1997).
Harding, T. M., Hefner-Gravink, A., Thumm, M. & Klionsky, D. J. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem. 271, 17621?17624 (1996).
Scott, S. V. et al. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl Acad. Sci. USA 93, 12304?12308 (1996).
Acknowledgements
Thanks to T. Noda and N. Mizushima for critical discussion and reading of this manuscript.
Author information
Authors and Affiliations
Glossary
- E1 ENZYME
-
An enzyme that activates the carboxy-terminal glycine of the small protein ubiquitin, or ubiquitin-like proteins, allowing them to form a high-energy bond to a specific cysteine residue of the E1.
- E2 ENZYME
-
An enzyme that accepts ubiquitin or a ubiquitin-like protein from an E1 and transfers it to the substrate, mostly using an E3 enzyme.
- NSF
-
N-ethylmaleimide-sensitive factor, an AAA-type ATPase essential for membrane fusion during vesicle transport.
- V-SNARE
-
A family of proteins on secretory vesicles. They form a complex with t-SNAREs on the target membrane during vesicle fusion.
Rights and permissions
About this article
Cite this article
Ohsumi, Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2, 211–216 (2001). https://doi.org/10.1038/35056522
Issue Date:
DOI: https://doi.org/10.1038/35056522
This article is cited by
-
Fluoride-Induced Mitochondrial Dysfunction and Approaches for Its Intervention
Biological Trace Element Research (2024)
-
Cd-Induced Autophagy Responses in Pakchoi as Revealed by Transcriptome Analysis
Plant Molecular Biology Reporter (2024)
-
Exon Sequence Analysis of the ATG5, ATG12, ATG9B Genes in Colorectal Cancer Patients During Radiotherapy
Indian Journal of Clinical Biochemistry (2024)
-
Research Progress on Histone Deacetylases Regulating Programmed Cell Death in Atherosclerosis
Journal of Cardiovascular Translational Research (2024)
-
Autophagy of Candida albicans cells after the action of earthworm Venetin-1 nanoparticle with protease inhibitor activity
Scientific Reports (2023)