Abstract
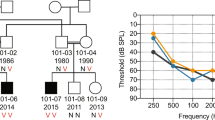
Nonsyndromic hearing impairment is one of the most heterogeneous hereditary conditions, with more than 40 loci mapped on the human genome1, however, only a limited number of genes implicated in hearing loss have been identified. We previously reported linkage to chromosome 7p15 for autosomal dominant hearing impairment segregating in an extended Dutch family2,3 (DFNA5). Here, we report a further refinement of the DFNA5 candidate region and the isolation of a gene from this region that is expressed in the cochlea. In intron 7 of this gene, we identified an insertion/deletion mutation that does not affect intron-exon boundaries, but deletes five G-triplets at the 3' end of the intron. The mutation co-segregated with deafness in the family and causes skipping of exon 8, resulting in premature termination of the open reading frame. As no physiological function could be assigned, the gene was designated DFNA5.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Van Camp, G., Willems, P.J. & Smith, R.J.H. Nonsyndromic hearing impairment: unparalleled heterogeneity. Am. J. Hum. Genet. 60, 758– 764 (1997).
Van Camp, G. et al. Localization of a gene for non-syndromic hearing loss (DFNA5) to chromosome 7p15. Hum. Mol. Genet. 4, 2159–2163 (1995).
Van Laer, L. et al. Refined mapping of a gene for autosomal dominant progressive sensorineural hearing loss (DFNA5) to a 2-cM region, and exclusion of a candidate gene that is expressed in the cochlea. Eur. J. Hum. Genet. 5, 397–405 (1997).
Kozak, M. Interpreting cDNA sequences: some insights from studies on translation. Mamm. Genome 7, 563–574 (1996).
Sirand-Pugnet, P., Durosay, P., Brody, E. & Marie, J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res. 23, 3501– 3507 (1995).
Cogan, J.D. et al. A novel mechanism of aberrant pre-mRNA splicing in humans. Hum. Mol. Genet. 6, 909– 912 (1997).
McCullough, A.J. & Berget, S.M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 17 , 4562–4571 (1997).
Thompson, D.A. & Weigel, R.J. Characterization of a gene that is inversely correlated with estrogen receptor expression (ICERE-1) in breast carcinomas. Eur. J. Biochem. 252, 169– 177 (1998).
Morton, N.E. Genetic epidemiology of hearing impairment. Ann. NY Acad. Sci. 630, 16–31 ( 1991).
Huizing, E.H., van Bolhuis, A.H. & Odenthal, D.W. Studies on progressive hereditary perceptive deafness in a family of 335 members. I. Genetical and general audiological results. Acta Otolaryngol. 61, 35– 41 (1966).
Huizing, E.H., van Bolhuis, A.H. & Odenthal, D.W. Studies on progressive hereditary perceptive deafness in a family of 335 members. II. Characteristic pattern of hearing deterioration. Acta Otolaryngol. 61, 161– 167 (1966).
Huizing, E.H., Odenthal, D.W. & van Bolhuis, A.H. Results of further studies on progressive hereditary sensorineural hearing loss. Audiology 12, 261–263 (1972).
Huizing, E.H., van den Wijngaart, W.S.I.M. & Verschuure, J. A follow-up study in a family with dominant progressive inner ear deafness. Acta Otolaryngol. 95 , 620–626 (1983).
van den Wijngaart, W.S.I.M., Verschuure, J., Brocaar, M.P. & Huizing, E.H. Follow-up study in a family with dominant progressive hereditary sensorineural hearing impairment. I. Analysis of hearing deterioration. Audiology 24, 233–240 ( 1985).
van den Wijngaart, W.S.I.M. et al. Follow-up study in a family with dominant progressive hereditary sensorineural hearing impairment. II. Clinical aspects. Audiology 24, 336–342 ( 1985).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997).
Xu, Y., Mural, R., Shah, M. & Uberbacher, E. Recognizing exons in genomic sequence using GRAIL II. Genet. Eng. 16, 241–253 (1994).
Zhang, M.Q. Identification of protein coding regions in the human genome by quadratic discriminant analysis. Proc. Natl Acad. Sci. USA 94 , 565–568 (1997).
Legan, P.K., Rau, A., Keen, J.N. & Richardson, G.P. The mouse tectorins. Modular matrix proteins of the inner ear homologous to components of the sperm-egg adhesion system. J. Biol. Chem. 272 , 8791–8801 (1997).
Wauters, J.G. et al. Regional mapping of a liver α-subunit gene of phosphorylase kinase (PHKA) to the distal region of human chromosome Xp. Cytogenet. Cell Genet. 60, 194–196 (1992).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 ( 1990).
Acknowledgements
The authors are grateful to all family members for their collaboration in this study. The adult mouse cochlear cDNA λ ZAP library was kindly provided by L. Hampton and J. Battey. We wish to thank F. Cremers for the Dutch control samples, B. Beverlo for the mouse telomere probes, E. Fransen for his suggestions concerning the nature of the mutation, E. Green for the BAC clones and S.-H. Correwyn for preparing figures. This study was supported by a grant from the University of Antwerp, and a grant from the Flemish Fonds voor Wetenschappelijk Onderzoek (FWO). G.V.C. holds a research position with the FWO. R.J.H.S. is supported in part by research grant R01 DC 03544.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Laer, L., Huizing, E., Verstreken, M. et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet 20, 194–197 (1998). https://doi.org/10.1038/2503
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/2503