Abstract
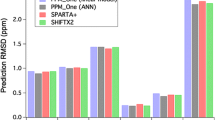
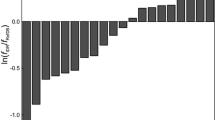
The importance of protein chemical shift values for the determination of three-dimensional protein structure has increased in recent years because of the large databases of protein structures with assigned chemical shift data. These databases have allowed the investigation of the quantitative relationship between chemical shift values obtained by liquid state NMR spectroscopy and the three-dimensional structure of proteins. A neural network was trained to predict the 1H, 13C, and 15N of proteins using their three-dimensional structure as well as experimental conditions as input parameters. It achieves root mean square deviations of 0.3 ppm for hydrogen, 1.3 ppm for carbon, and 2.6 ppm for nitrogen chemical shifts. The model reflects important influences of the covalent structure as well as of the conformation not only for backbone atoms (as, e.g., the chemical shift index) but also for side-chain nuclei. For protein models with a RMSD smaller than 5 Å a correlation of the RMSD and the r.m.s. deviation between the predicted and the experimental chemical shift is obtained. Thus the method has the potential to not only support the assignment process of proteins but also help with the validation and the refinement of three-dimensional structural proposals. It is freely available for academic users at the PROSHIFT server: www.jens-meiler.de/proshift.html
Similar content being viewed by others
References
Bonneau, R., Tsai, J., Ruczinski, I., Chivian, D., Rohl, C., Strauss, C. E. M. and Baker, D. (2001) Proteins, 45(Suppl.), 119-126.
Braun, D., Wider, G. and Wuethrich, K. (1994) J. Am. Chem. Soc., 116, 8466-8469.
Chandonia, J.-M. and Karplus, M. (1999) Proteins Struct. Funct. Genet., 35, 293-306.
Choy, W.Y., Sanctuary, B.C. and Zhu, G. (1997) J. Chem. Inf. Comput. Sci., 37, 1086-1094.
Clouser, D.L. and Jurs, P.C. (1996) Anal. Chim. Acta, 321, 127-135.
Cornilescu, G., Delaglio, F. and Bax, A. (1999) J. Biomol. NMR, 13, 289-302.
Gronwald, W., Boyko, R.F., Sonnichsen, F.D., Wishart, D.S. and Sykes, B.D. (1997) J. Biomol. NMR, 10, 165-179.
Ivanciuc, O., Rabine, J.P., Cabrol-Bass, D., Panaye, A. and Doucet, J.-P. (1996) J. Chem. Inf. Comput. Sci., 36, 644-653.
Iwadate, M., Asakura, T. and Williamson, M.P. (1999) J. Biomol. NMR, 13, 199-211.
Jones, D.T. (1999) J. Mol. Biol., 292, 195-202.
Kneller, D.G., Cohen, F.E. and Langridge, R. (1990) J. Mol. Biol., 214, 171-182.
Kvasnicka, V., Sklenak, S. and Pospichal, J. (1992) J. Chem. Inf. Comput. Sci., 32, 742-747.
Le, H. and Oldfield, E. (1994) J. Biomol. NMR, 4, 341-348.
Luman, N.R., King, M.P. and Augspurger, J.D. (2001) J. Comput. Chem., 22, 366-372.
Meiler, J. (1996-2002) www.jens-meiler.de
Meiler, J. (2002a) www.jens-meiler.de/jufo.html
Meiler, J. (2002b) www.jens-meiler.de/proshift.html
Meiler, J. and Will, M. (2001) J. Chem. Inf. Comput. Sci., 41, 1535-1546.
Meiler, J., Maier, W., Will, M. and Meusinger, R. (2002) J. Magn. Reson., 157, 242-252.
Meiler, J., Müller, M., Zeidler, A. and Schmäschke, F. (2001) J. Mol. Model., 7, 360-369.
Meiler, J., Will, M. and Meusinger, R. (2000) J. Chem. Inf. Comput. Sci., 40, 1169-1176.
Meusinger, R. and Moros, R. (1995) In Software - Entwicklung in der Chemie, Vol. 10, Gasteiger, J., Ed. Gesellschaft Deutscher Chemiker, Frankfurt am Main, pp. 209-216.
Oldfield, E. (1995) J. Biomol. NMR, 5, 217-225.
Osapay, K. and Case, D.A. (1991) J. Am. Chem. Soc., 113, 9436-9444.
Pearson, J.G., Le, H., Sanders, L.K., Godbout, N., Havlin, R.H. and Oldfield, E. (1997) J. Am. Chem. Soc., 119, 11941-11950.
Petersen, T.N., Lundegaard, C., Nielsen, M., Bohr, H., Bohr, J., Brunak, S., Gippert, G.P. and Lund, O. (2000) Proteins Struct. Funct. Genet., 41, 17-20.
Pons, J.L. and Delsuc, M.A. (1999) J. Biomol. NMR, 15, 15-26.
Qian, N. and Sejnowski, T.J. (1988) J. Mol. Biol., 202, 865-884.
Ramirez, B.E., Voloshin, O.N., Camerini-Otero, R.D. and Bax, A. (2000) Protein Sci., 9, 2161.
Robien, W. (1998) Nachr. Chem. Tech. Lab., 46, 74-77.
Rohl, C. and Baker, D. (2002) J. Am. Chem. Soc., 124, 2723-2729.
Rost, B. (1996) Meth. Enzymol., 266, 525-539.
Rost, B. and Sander, C. (1993) J. Mol. Biol., 232, 584-599.
Rost, B., Sander, C. and Schneider, R. (1994) J. Mol. Biol., 235, 13-26.
Salamov, A.A. and Solovyev, V.V. (1997) J. Mol. Biol., 268, 31-36.
Spera, S. and Bax, A. (1991) J. Am. Chem. Soc., 113, 5490-5492.
Stolorz, P., Lapedes, A. and Xia, Y. (1992) J. Mol. Biol., 225, 363-377.
Thomas, S. and Kleinpeter, E. (1995) J. Prakt. Chem./Chem.-Ztg., 337, 504-507.
Wang, Y. and Jardetzky, O. (2002) Protein Sci., 11, 852-861.
Wishart, D.S. and Sykes, B.D. (1994) J. Biomol. NMR, 4, 171-180.
Wishart, D.S., Bigam, C.G., Holm, A., Hodges, R.S. and Sykes, B.D. (1995) J. Biomol. NMR, 5, 67-81.
Wishart, D.S., Sykes, B.D. and Richards, F.M. (1992) Biochemistry, 31, 1647-1651.
Wishart, D.S., Watson, M.S., Boyko, R.F. and Sykes, B.D. (1997) J. Biomol. NMR, 10, 329-336.
Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (1HNMR Shifts of Amino Acids), John Wiley & Sons, New York, Chichester, Brisbane, Toronto, Singapore.
Xu, X.-P. and Case, D.A. (2001) J. Biomol. NMR, 21, 321-333.
Zupan, J. and Gasteiger, J. (1993) Neural Networks for Chemists, VCH Verlagsgesellschaft mbH, Weinheim.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meiler, J. PROSHIFT: Protein chemical shift prediction using artificial neural networks. J Biomol NMR 26, 25–37 (2003). https://doi.org/10.1023/A:1023060720156
Issue Date:
DOI: https://doi.org/10.1023/A:1023060720156