INTRODUCTION
Norovirus (NoV) is the most common cause of outbreaks of acute gastroenteritis (AGE) worldwide [Reference Fankhauser1–Reference Svraka5]. In The Netherlands, 78% of the 941 reported gastroenteritis outbreaks during 1994–2005 were caused by NoV infection [Reference Svraka5]. In Oregon, 50% of the 1047 AGE outbreaks reported during 1999–2008 were caused by NoV infection [Oregon Public Health Division (OPHD), unpublished data]. NoV outbreaks can occur in many settings, but they disproportionately affect closed or semi-closed settings including long-term care facilities (LTCFs), hospitals, and cruise ships [6]. In Switzerland, 59% of NoV outbreaks during 2001–2003 occurred in nursing homes and hospitals [Reference Fretz7]. In Oregon, 65% of NoV outbreaks occurred in LTCFs during 1999–2008 (OPHD, unpublished data). Although NoV infection is often self-limited, it can cause severe dehydration, hospitalization, and even death, especially in older persons [Reference Harris8–Reference Mattner10]. According to the U.S. Census Bureau's projection, persons aged ⩾65 years will account for 25% of the US general population by 2050, and a substantial number of these persons will need long-term care services [11]. According to the National Nursing Home Survey, the number of beds in nursing homes increased by 47% from 1973 to 2004 [12]. A better understanding of outbreaks in LTCFs might help reduce NoV-related infections, hospitalizations, and deaths in older populations and minimize NoV-related economic costs [Reference Johnston13, Reference Zingg14].
No comprehensive surveillance system for NoV infections exists in the USA. Individual infections are typically not reportable. Outbreaks often are reportable, but prior to 2008, only foodborne outbreak data have been actively collected at the national level, which may only represent a limited number of NoV outbreaks [Reference van Duynhoven15]. Outbreaks resulting from person-to-person transmission, which is usually the primary transmission mode for AGE outbreaks in LTCFs, are often incompletely investigated and rarely tested for NoV in many states. Consequently, epidemiological characteristics of NoV outbreaks in LTCFs in the USA are not well documented.
NoV is genetically heterogeneous [Reference Zheng16]. Strains from three of the five genogroups have been linked to human infections and can be divided into ⩾26 genetic clusters [Reference Zheng16]. The majority of outbreaks in certain countries and settings have been caused by NoV genogroup II genotype 4 (GII.4) strains [Reference Lopman4, Reference Blanton17]. In The Netherlands, GII.4 strains accounted for 89% of the NoV outbreaks during 2004–2005 [Reference Siebenga18]. In Maryland (USA), 14 (70%) of the 20 nursing home-related outbreaks reported during the 1987–1988 winter season were caused by NoV GII.4 strains [Reference Green19]. During the past two decades, emerging variants of GII.4 strains have been linked to increases in the number of AGE outbreaks worldwide every 3–4 years [Reference Kageyama2–Reference Lopman4, Reference Blanton17, Reference Bull20, Reference Siebenga21]. One such emergent GII.4 variant strain, Farmington Hills/2002, which emerged in 2002, has been linked to a slightly higher prevalence of diarrhoea and a greater attack rate [Reference Blanton17, Reference Widdowson22]. These studies highlighted the need to better understand the association between NoV GII.4 strains and the clinical manifestations of NoV infection, as well as the molecular evolution of GII.4 strains with time. In this study, we used systematically collected outbreak surveillance data to investigate the epidemiological and genetic characteristics of NoV outbreaks in Oregon LTCFs during 2003–2006.
MATERIALS AND METHODS
Data collection
In Oregon, all outbreaks are reportable by law. Local (county) health departments usually take the lead in investigating AGE outbreaks under the supervision of one epidemiologist in OPHD. For AGE outbreaks in LTCFs, local health departments often delegate data-collection responsibilities to nurses in the facilities. Standardized OPHD protocols have been used for investigating AGE outbreaks in all LTCFs in Oregon since 2003. Individual case data (i.e. demographics, resident or staff status, clinical symptoms and onset time) are collected on a standardized form by facility nurses on a daily basis based on each individual's self-report. If a point source of infection is suggested by the epidemic curve and other epidemiological evidence, an investigation of the source of exposure is warranted. If person-to-person transmission is indicated, no investigation on source of exposure is conducted. Summary data for each LTCF outbreak, including first and last onset dates, primary transmission mode, total number of cases, and number and type of specimens collected, are recorded on a separate form. All outbreak and case data are entered into an electronic outbreak surveillance database at OPHD. We solicited stool specimens from a convenience sample of at least three ill and symptomatic persons in each outbreak for testing.
Study measures and definitions
LTCFs were defined as institutions licensed by the Oregon Department of Human Services (ODHS) and provide a range of services for persons who have limitations in daily activities. A list of LTCFs was obtained from ODHS in mid-2005. The number of LTCFs fluctuated only slightly during 2003–2006, and the number and characteristics of the LTCFs in mid-2005 were typical for the study period. We categorized LTCFs as ‘nursing facilities’ if they provided 24-h nursing care or ‘non-nursing’ facilities otherwise. LTCFs were also categorized as large (⩾90 beds) or small (<90 beds). Individual cases were categorized as being related to employees or residents.
An AGE outbreak was defined as an outbreak in which the initial cluster included at least three persons in the same LTCF who were experiencing acute onset of diarrhoea or vomiting within a 4-day period. To be classified as an aetiologically confirmed outbreak, stool specimens from at least two persons must have been tested positive for the same pathogen and no other pathogens [Reference Widdowson22]. Confirmed NoV cases had a polymerase chain reaction (PCR)-positive stool specimen; presumptive NoV cases had acute vomiting or diarrhoea in the context of a confirmed NoV outbreak without laboratory confirmation. Both confirmed and presumptive NoV cases were included in the analyses. NoV outbreak-associated hospitalizations were defined as any overnight stay in a hospital resulting from AGE in a LTCF resident with confirmed or presumptive NoV infection during a confirmed NoV outbreak. NoV outbreak-related deaths referred to any death occurring in a LTCF resident with confirmed or presumptive NoV infection during a confirmed NoV outbreak.
The annual attack rate for NoV infection in LTCF residents during the 4-year period was calculated by dividing the total number of resident cases by the total number of residents in LTCFs, and dividing by four. The number of LTCF residents was estimated by 80% of the total number of beds in all facilities combined (i.e. assuming an average occupancy of 80% based on findings from a survey of a representative sample of LTCFs in Oregon conducted in October, 2007). The case-hospitalization rate was calculated by dividing the reported number of resident hospitalizations by the number of resident cases. The case-fatality rate was calculated by dividing the number of resident deaths by the number of resident cases.
Laboratory testing and genotyping
For each outbreak, if a viral pathogen was suspected (e.g. based on incubation period, duration of illness, and symptoms), stool specimens were first tested for NoV by reverse transcriptase–polymerase chain reaction (RT–PCR) at the OPHD laboratory. If all specimens were negative for NoV, the specimens were sent to the Minnesota Public Health Laboratory for further testing for other viruses. Specimens that tested positive for NoV during 2003–2006 were sent to the National Calicivirus Libratory for genotyping. If a bacterial pathogen was suspected, routine bacterial culture was performed according to standard procedures. For NoV testing, viral RNA was extracted from a 10% stool suspension by using either QIAamp Viral RNA Mini kit (Qiagen, USA) or MagAttract® Viral RNA M48 kit (Qiagen 955235) on the Qiagen M48 BioRobot® automated extractor. Before 2006, NoV GI and GII detection was performed by using Titan One Tube RT–PCR kit (Roche, USA) and oligonucleotide primers targeting region B of ORF1 (MON431, 432, 433 and 434) [Reference Trujillo23, Reference Kageyama24]. Cycling conditions included reverse transcription at 42°C for 10 min, activation of Taq polymerase at 94°C for 3 min, followed by 40 cycles at 94°C for 30 s, 50°C for 1·5 min, and 60°C for 30 s. RT–PCR products were separated on a 3·5% agarose gel and visually evaluated by ethidium bromide staining of a 213-bp product. Since 2006, NoV GI and GII were detected by TaqMan® (Applied Biosystems, USA) real-time (q)RT–PCR [Reference Trujillo23, Reference Kageyama24] optimized for an ABI 7500 platform. The final reaction mixture included 400 nm of oligonucleotide primers and 150 nm of probe in a Quantitect® Probe RT–PCR Mix (Qiagen). Cycling conditions included reverse transcription at 50°C for 30 min, Taq activation at 95°C for 10 min, and 45 cycles of 95°C for 15 s and 60°C for 1 min.
For NoV genotyping, viral nucleic acids were extracted from clarified 10% stool suspensions by using the KingFisher instrument and MagMAXTM-96 Viral RNA Isolation kit (Ambion, USA) according to the manufacturer's instructions. Genotyping was performed by amplifying partial capsid regions (region C [Reference Kojima25], region D [Reference Vinje, Hamidjaja and Sobsey26]) of the genome by RT–PCR with the One-Step® RT–PCR Kit (Qiagen). After cycle sequencing with the BigDye® Terminator Cycle Sequencing Reaction kit (Applied Biosystems), samples were analysed on an ABI Prism® 3130xl Genetic Analyzer (Applied Biosystems). Sequences were edited by using Sequencher® (Gene Core Corporation, USA), and multiple alignments were created by using Clustal W and imported into TreeCon [Reference Van de Peer and De Wachter27] to generate phylogenetic trees. NoV prototype strains [Reference Zheng16], as well as GII.4 variant strains (Farmington Hills/2002 AY502023, Hunter/2004 DQ078794, Minerva/2006b EU078417, Terneuzen/2006a EF126964), were included as reference strains.
Statistical analysis
SAS® version 9.1.3 (SAS, USA) was used for analysis. We assessed the associations between facility type and size, and risk and duration of NoV outbreaks using multivariable binomial and linear regression models, respectively. We used the Mantel–Haenszel test to estimate the adjusted effect of facility size and type on the risk for experiencing repeated NoV outbreaks. We used bivariate binomial regression to estimate the associations between genotype and prevalence of vomiting, diarrhoea, physician examination, and hospitalization in case residents. Duration of symptoms between patients with GII.4 infections and other genotypes was compared by Wilcoxon signed ranks test. The χ2 test for trend was used to assess change in proportion of outbreaks caused by GII.4 strains during the study period. Statistical significance level was set at α=0·05.
RESULTS
Epidemiological characteristics of NoV outbreaks
There were 533 LTCFs licensed in Oregon as of mid-2005, including 126 (24%) nursing facilities, of which 63 (50%) were large facilities. Of the 407 non-nursing facilities, only 64 (16%) were large facilities (χ2=67·2, P<0·0001).
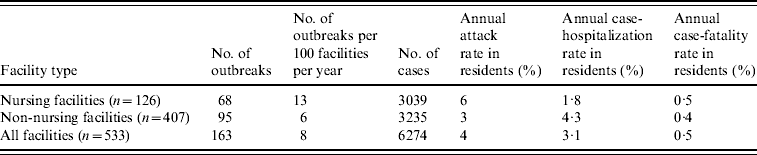
During the 4-year study period, 234 LTCF outbreaks were reported, of which 165 (71%) were aetiologically confirmed; 29 (13%) had one specimen tested positive for NoV; 18 (7%) had all specimens tested negative for NoV; and 22 (9%) had no specimens available for testing. Of the 165 aetiologically confirmed outbreaks, 163 (99%) comprising at least 6274 cases (541 confirmed and 5733 presumptive) were caused by NoV and two were caused by Salmonella. The average age of resident cases was 82·6 years (range 18–106 years). The median number of cases per outbreak was 43 (range 5–154) for outbreaks in nursing facilities, 28 (range 3–117) for non-nursing facilities, 48 (range 7–154) for large facilities, and 32 (range 3–117) for small facilities. Of all confirmed NoV outbreaks, primary transmission mode was reported as person-to-person for 94%, foodborne for 2·5%, and undetermined for 3·5%. The annual attack rate for NoV outbreaks was 13% in nursing facilities vs. 6% in non-nursing facilities. The annual attack rate for NoV infections in LTCF residents was 6% in nursing facilities and 3% in non-nursing facilities. Of NoV cases in LTCF residents, 180 cases were hospitalized and 20 cases died during the outbreak. The case-hospitalization and case-fatality rates in resident cases were 1·8% and 0·5% in nursing facilities, and 4·3% and 0·4% in non-nursing facilities (Table 1). No employee cases were hospitalized or died.
Table 1. Epidemiological characteristics of confirmed norovirus outbreaks in long-term care facilities (LTCFs) by facility type, Oregon, USA, 2003–2006

The number of facilities was obtained from a list of licensed LTCFs in 2005 provided by the Oregon Department of Human Services Division of Services for Seniors and the Disabled. For the calculation of risk in residents, the total number of LTCF residents was estimated by multiplying the total number of beds with the estimated occupancy rate (80%).
Facility-level risk factors for NoV outbreaks
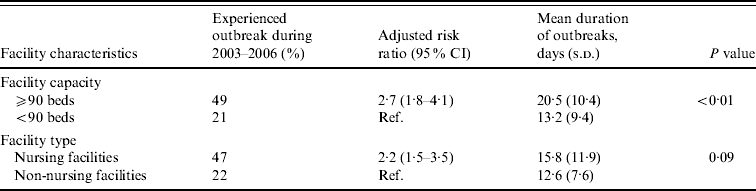
Large facilities were 2·7 times more likely to report NoV outbreaks than small facilities after adjusting for facility type (Table 2). Outbreaks persisted an average of 7 days longer in large compared to small facilities (mean 20 days vs. 13 days, P<0·01). Nursing facilities were 2·2 times more likely to report NoV outbreaks than non-nursing facilities after adjusting for facility size.
Table 2. Association between facility features and risk and mean duration of norovirus outbreaks in long-term care facilities, Oregon, USA, 2003–2006

CI, Confidence interval; s.d., standard deviation.
Facility capacity and type were adjusted for each other.
Multiple outbreaks at the same facility
Eleven nursing facilities (9%) and eight non-nursing facilities (2%) reported two or three outbreaks during the study period. The median time interval between recurring outbreaks was 14 months (range 2–45 months). After adjusting for facility size, nursing facilities were 2·3 times more likely [95% confidence interval (CI) 1·0–4·9] to report multiple outbreaks than non-nursing facilities. After adjusting for facility type, large facilities were 5·9 times (95% CI 2·4–14·8) more likely to report multiple outbreaks than small facilities. Of the 19 LTCFs experiencing repeated outbreaks, in five (26%), subsequent outbreaks were caused by a NoV strain identical to the initial outbreak; in 11 (58%), subsequent outbreaks were caused by NoV strains different from previous outbreaks; and in three (16%), outbreak strain was not determined for at least one of the repeated outbreaks. At least 86 persons (20 employees, 66 residents) were infected in multiple outbreaks.
Effect of NoV genotype on severity and duration of NoV infection
Genotype was determined for 129 (79%) of 163 NoV outbreaks. Overall, strains belonging to eight NoV genotypes (GI.1, GI.4, GI.6, GII.3, GII.4, GII.5, GII.6, GII.10) were detected in LTCFs during the study period. GII.4 strains accounted for 108 (84%) of the NoV outbreaks with known genotype. No statistically significant differences were observed in the risks for vomiting, diarrhoea, being examined by a physician, or being hospitalized between cases with GII.4 infections and those with other genotype infections (Table 3). The median duration of illness was 33 h (range 1·5–216 h) for cases caused by GII.4 strains and 24 h (range 2–168 h) for those caused by other NoV genotypes (P<0·001).
Table 3. Association between norovirus genotype and clinical outcomes in case persons involved in norovirus outbreaks in long-term care facilities, Oregon, USA, 2003–2006

RR, Relative risk; CI, confidence interval.
For all comparisons, Non-GII.4 group was the reference group.
* A comparison between case persons with GII.4 infection and non-GII.4 infection.
Seasonality and genotype trends
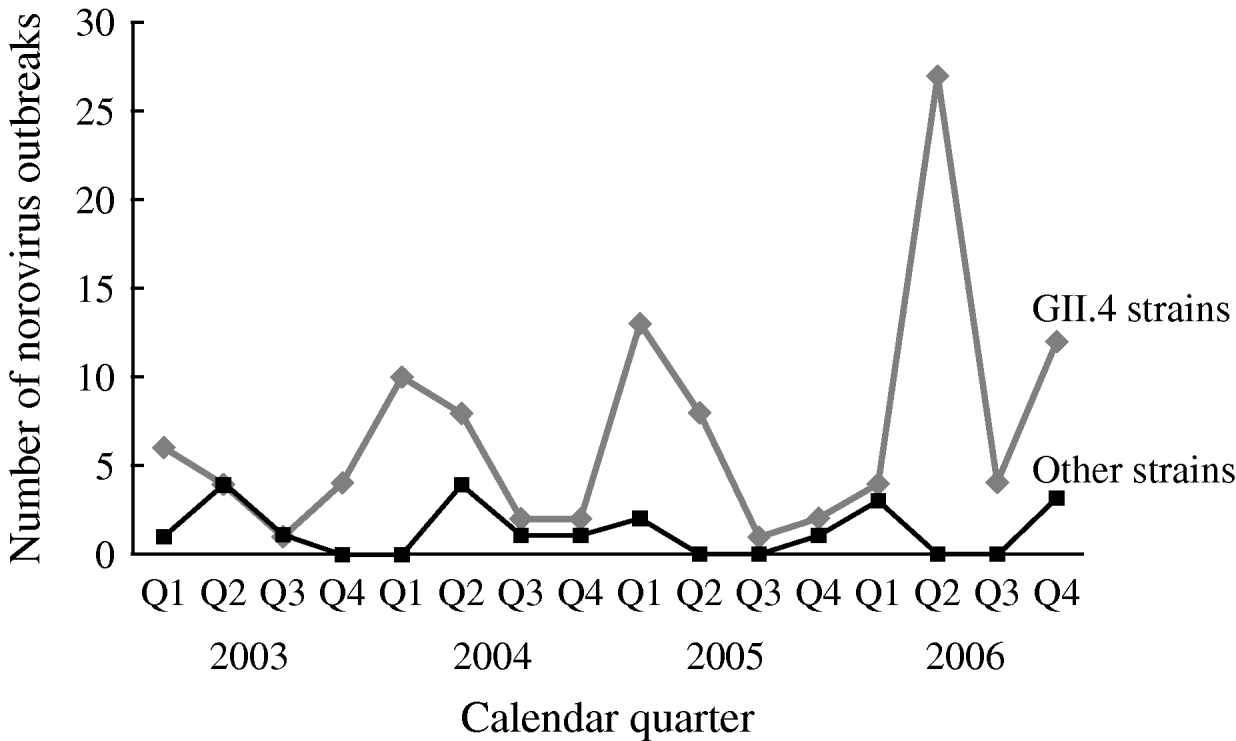
The number of reported NoV outbreaks increased from 31 in 2003 to 67 in 2006. The proportion of outbreaks caused by GII.4 strains increased from 71% in 2003 to 89% in 2006 (P trend=0·056). GII.4 outbreaks peaked in winter and spring, with a dramatic increase in the second quarter of 2006. There was no obvious seasonality observed for non-GII.4 strain outbreaks (Fig. 1).

Fig. 1. Time trend and seasonality of GII.4 and non-GII.4 norovirus outbreaks in long-term care facilities, Oregon, USA, 2003–2006 (n=129).
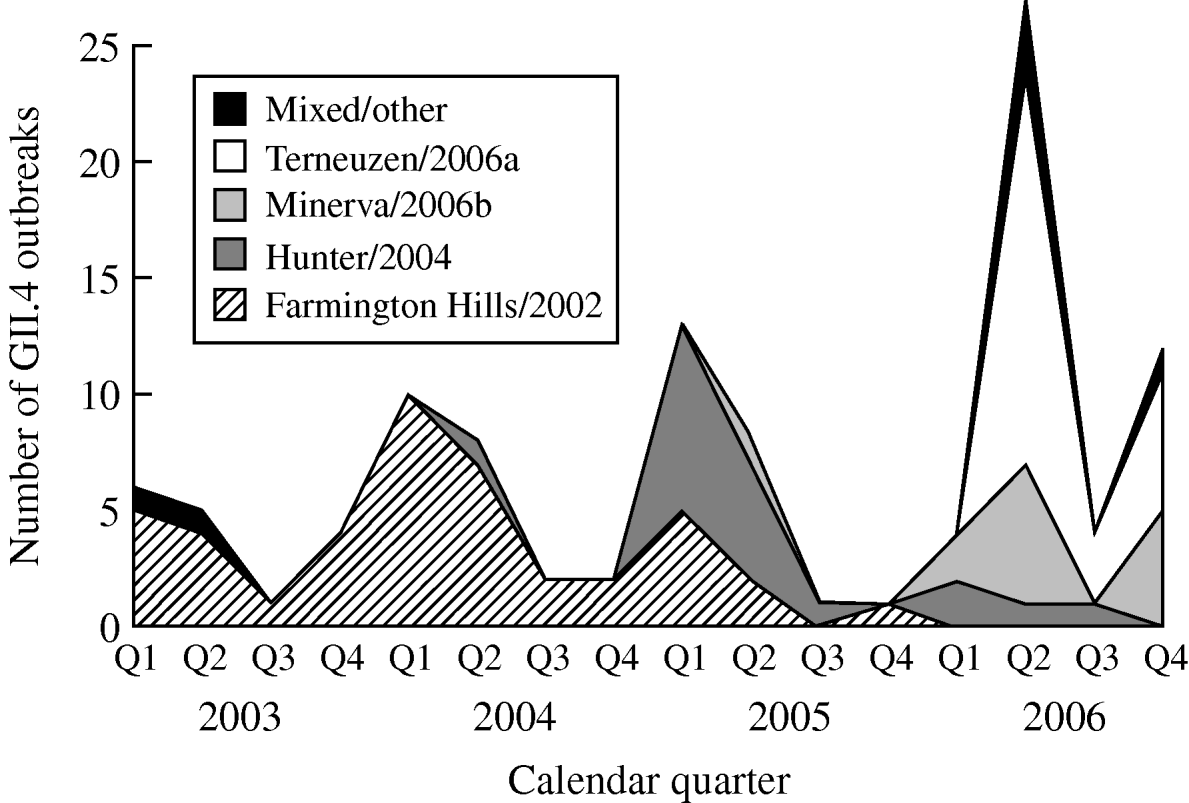
From 2003 to 2006, GII.4 Farmington Hills/2002 strains were steadily replaced by other GII.4 strains. Farmington Hills/2002 strains accounted for 94% of the GII.4 outbreaks in 2003, 95% in 2004, 35% in 2005, and 2% in 2006. Emerging GII.4 strains Hunter/2004, Minerva/2006b, and Terneuzen/2006a first appeared in the second quarter of 2004, 2005, and 2006, respectively (Fig. 2).

Fig. 2. Number of outbreaks due to different norovirus GII.4 variant strains in long-term care facilities by calendar quarter and year, Oregon, USA, 2003–2006 (n=108). Q1, First quarter (January–March); Q2, second quarter (April–June); Q3, third quarter (July–September); Q4, fourth quarter (October–December). In the ‘Mixed/other’ category, the one in the first quarter of 2003 was a GII.4 strain that was different from any named strains; the one in the second quarter of 2003 was a mix of GII.4 Farmington Hills/2002 and GII.3; the three in the second quarter of 2006 were a mix of GII.4 Minerva/2006b and Terneuzen/2006a; and the one in the fourth quarter of 2006 was a mix of GII.4 Farmington Hills/2002 and Minerva/2006b.
DISCUSSION
The majority of aetiologically confirmed outbreaks of AGE in Oregon LTCFs are caused by NoV. This indicates that when an AGE outbreak occurs in a LTCF, it is reasonable to assume NoV as the most probable cause until proven otherwise, and to promptly implement appropriate control measures.
NoV causes substantial morbidity and mortality in LTCFs, with at least 8% of facilities afflicted each year. Observed case-hospitalization rate (3·1%) and case-fatality rate (0·5%) in resident cases were somewhat lower than rates reported in Israeli nursing homes (10·2% and 2·0%, respectively) and were also different from rates reported in nursing homes in The Netherlands (0·5% and 1·6%, respectively) [Reference Calderon-Margalit28, Reference Friesema29], which may reflect chance or differences in the study populations or outbreak and case definitions. The six facilities included in the Israeli study were all nursing homes, with residents that may be less robust than our LTCF population [Reference Calderon-Margalit28]. The severity of NoV infections in generally healthier populations with less pre-existing conditions is clearly much less. The NoV infection-related hospitalization rate in cases from the general population was reported as 0·3% in Britain [Reference Lopman30], and 1% in the USA [Reference Widdowson31].
Larger facilities and those that offer more intensive nursing care may be at higher risk for having NoV outbreaks than smaller facilities and those without intensive nursing services, although we can not rule out reporting bias as a source of the observed differences. In our study, facility size seemed to have a greater effect on risk for occurrence and recurrence of NoV outbreaks than facility type. This is consistent with findings from studies in The Netherlands and Japan [Reference Yamamoto32, Reference Friesema33]. NoV is highly contagious, and after the virus is introduced into a LTCF, especially a large facility, an outbreak is almost unavoidable if the facility does not have thorough hygienic and infection-control practices.
Certain LTCFs experienced repeated outbreaks: sometimes with identical strains, and sometimes with different strains. A number of individuals were also documented to have recurrent infections. This is consistent with prior findings from human volunteer studies indicating that no long-term protective immunity could be developed after exposure to NoV infections [Reference Matsui and Greenberg34, Reference Parrino35]. This poses a great challenge for vaccine development and infection control.
The number of NoV outbreaks more than doubled from 2005 to 2006. This might be associated with the emergence of two novel GII.4 variant strains – GII.4 Minerva/2006b and GII.4 Terneuzen/2006a. A similar increase of the same GII.4 strains – albeit not as dramatic – was noted in Europe [Reference Kroneman36]. We found that the dominant GII.4 strain in 2003 (Farmington Hills) was gradually replaced by other emerging GII.4 strains: Hunter/2004 in 2004, Hunter/2004 and Minerva/2006b in 2005, and Minerva/2006b and Terneuzen/2006a in 2006. These findings are consistent with prior research indicating that GII.4 viruses evolve stepwise by antigenic drift and escape from herd immunity in the population, and that new GII.4 variants constantly emerge and displace previously circulating strains [Reference Siebenga37–Reference Cannon39].
Outbreaks attributable to GII.4 strains, unlike those resulting from non-GII.4 strains, appear to have a distinct seasonal pattern, peaking in winter or spring [Reference Lopman30, Reference Lopman, Brown and Koopmans40, Reference Mounts41]. The dramatic increase in NoV GII.4 outbreaks in 2006 mainly occurred during spring. We also noted that NoV GII.4 strains were associated with prolonged illness. With prolonged duration of illness, cases might shed NoV in greater numbers, as indicated in a recent volunteer study [Reference Atmar42], thereby increasing the probability of transmission. Friesema et al. found in a sample of nursing homes in The Netherlands that NoV GII.4 infection was associated with higher vomiting prevalence in residents but not in staff compared to non-GII.4 infection, No association was found between genotype and duration of illness in either staff or residents [Reference Friesema29]. The inconsistency between our findings and theirs implies the need for further research studying the clinical characteristics of NoV GII.4 infection. To reduce NoV-related disease burden, GII.4 strains need to be targeted by future NoV research concerning effective infection-control strategies.
The current study has at least three limitations. First, although all outbreaks are reportable by law in Oregon, underreporting is unavoidable. Thus, our estimates of the public health impact of these outbreaks represent a lower bound. Second, because residents in nursing facilities tend to be under closer observation of nurses than residents in non-nursing facilities, outbreaks in the former are probably more likely to be identified and reported. Third, because we requested two or more positive specimens to confirm a NoV outbreak, the proportion of outbreaks caused by NoV might have been underestimated. The 13% of outbreaks with only one specimen tested positive for NoV and some of the 7% of outbreaks with no specimens collected could have been attributed to NoV if sufficient specimens had been tested. Therefore, the impact of NoV on the LTCF population might be underestimated.
Our data confirm that NoV is the most likely cause of outbreaks of AGE in LTCFs, with GII.4 viruses as the most prevalent genotype. These GII.4 strains change over time, and these shifts sometimes herald changes in the incidence of outbreaks. For example, the increase of NoV outbreaks in 2006 was directly correlated with the emergence of two new GII.4 variant strains. Overall, outbreaks caused by GII.4 viruses were associated with longer duration of illness in LTCF residents. The susceptibility of the growing number of LTCF residents, the highly infectious and persistent nature of NoV, and the emergence of novel GII.4 strains underscore the need for more effective infection-control strategies in LTCFs. Future studies should focus on assessing the efficacy of current, and the feasibility of improved, infection-control measures in LTCFs.
ACKNOWLEDGEMENTS
The authors thank Tasha M. Poissant and John Rodakowski for their assistance with data management; Christianne Biggs, LaDonna Grenz, and Marjorie Yungclas at the Oregon State Public Health Laboratory and Kara Williams at CDC for technical assistance; Elaine Young at the Oregon Department of Human Services, Division of Services for Seniors and the Disabled, for providing information regarding long-term care facility regulations; and the communicable disease nurses and environmental health specialists at the Oregon local health departments who investigated NoV outbreaks and provided data for this report. We also thank Dr Sheryl Lyss at CDC for her helpful comments and suggestions. Thanks are also due to the Emerging Infectious Disease programme at the Centers for Disease Control and Prevention for their funding to Oregon.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC). This article did receive clearance through the appropriate channels at CDC before submission.
DECLARATION OF INTEREST
None.