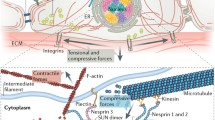
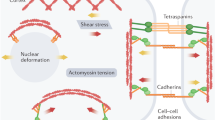
Abstract
Mechanical forces drive and modulate a wide variety of processes in eukaryotic cells including those occurring in the nucleus. Relevantly, forces are fundamental during development since they guide lineage specifications of embryonic stem cells. A sophisticated macromolecular machinery transduces mechanical stimuli received at the cell surface into a biochemical output; a key component in this mechanical communication is the cytoskeleton, a complex network of biofilaments in constant remodeling that links the cell membrane to the nuclear envelope. Recent evidence highlights that forces transmitted through the cytoskeleton directly affect the organization of chromatin and the accessibility of transcription-related molecules to their targets in the DNA. Consequently, mechanical forces can directly modulate transcription and change gene expression programs. Here, we will revise the biophysical toolbox involved in the mechanical communication with the cell nucleus and discuss how mechanical forces impact on the organization of this organelle and more specifically, on transcription. We will also discuss how live-cell fluorescence imaging is producing exquisite information to understand the mechanical response of cells and to quantify the landscape of interactions of transcription factors with chromatin in embryonic stem cells. These studies are building new biophysical insights that could be fundamental to achieve the goal of manipulating forces to guide cell differentiation in culture systems.



Similar content being viewed by others
References
Amendola M, van Steensel B (2015) Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep 16(5):610–617. https://doi.org/10.15252/embr.201439789
Block J, Schroeder V, Pawelzyk P, Willenbacher N, Koster S (2015) Physical properties of cytoplasmic intermediate filaments. Biochim Biophys Acta 1853(11):3053–3064. https://doi.org/10.1016/j.bbamcr.2015.05.009
Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K (1999) Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet 21(3):285–288. https://doi.org/10.1038/6799
Boraas LC, Guidry JB, Pineda ET, Ahsan T (2016) Cytoskeletal expression and remodeling in pluripotent stem cells. PLoS ONE 11(1):e0145084. https://doi.org/10.1371/journal.pone.0145084
Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, Weitz DA (2006) Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol 173(5):733–741. https://doi.org/10.1083/jcb.200601060
Brangwynne CP, MacKintosh FC, Weitz DA (2007) Force fluctuations and polymerization dynamics of intracellular microtubules. Proc Natl Acad Sci U S A 104(41):16128–16133. https://doi.org/10.1073/pnas.0703094104
Briand N, Collas P (2020) Lamina-associated domains: peripheral matters and internal affairs. Genome Biol 21(1):85. https://doi.org/10.1186/s13059-020-02003-5
Broers JL, Peeters EA, Kuijpers HJ, Endert J, Bouten CV, Oomens CW, Baaijens FP, Ramaekers FC (2004) Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum Mol Genet 13(21):2567–2580. https://doi.org/10.1093/hmg/ddh295
Burke B, Roux KJ (2009) Nuclei take a position: managing nuclear location. Dev Cell 17(5):587–597. https://doi.org/10.1016/j.devcel.2009.10.018
Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117(4):427–439. https://doi.org/10.1016/S0092-8674(04)00448-9
Charrier EE, Janmey PA (2016) Mechanical properties of intermediate filament proteins. Methods Enzymol 568:35–57. https://doi.org/10.1016/bs.mie.2015.09.009
Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB (2006) Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells 24(1):177–185. https://doi.org/10.1634/stemcells.2004-0159
Cosentino MS, Oses C, Vazquez Echegaray C, Solari C, Waisman A, Alvarez Y, Petrone MV, Francia M, Schultz M, Sevlever G, Miriuka S, Levi V, Guberman A (2019) Kat6b modulates Oct4 and Nanog binding to chromatin in embryonic stem cells and is required for efficient neural differentiation. J Mol Biol 431(6):1148–1159. https://doi.org/10.1016/j.jmb.2019.02.012
Dahal L, Walther N, Tjian R, Darzacq X, Graham TGW (2023) Singlemolecule tracking (SMT): a window into live-cell transcription biochemistry. Biochem Soc Trans 51(2):557–569. https://doi.org/10.1042/BST20221242
Dahl KN, Kahn SM, Wilson KL, Discher DE (2004) The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci 117(20):4779–4786. https://doi.org/10.1242/jcs.01357
Danchenko M, Csaderova L, Fournier PE, Sekeyova Z (2019) Optimized fixation of actin filaments for improved indirect immunofluorescence staining of rickettsiae. BMC Res Notes 12(1):657. https://doi.org/10.1186/s13104-019-4699-9
David BG, Fujita H, Yasuda K, Okamoto K, Panina Y, Ichinose J, Sato O, Horie M, Ichimura T, Okada Y, Watanabe TM (2019) Linking substrate and nucleus via actin cytoskeleton in pluripotency maintenance of mouse embryonic stem cells. Stem Cell Res 41:101614. https://doi.org/10.1016/j.scr.2019.101614
De Belly H, Paluch EK, Chalut KJ (2022) Interplay between mechanics and signalling in regulating cell fate. Nat Rev Mol Cell Biol 23(7):465–480. https://doi.org/10.1038/s41580-022-00472-z
De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, Levy N (2003) Lamin a truncation in Hutchinson-Gilford progeria. Science 300(5628):2055. https://doi.org/10.1126/science.1084125
de Leeuw R, Gruenbaum Y, Medalia O (2018) Nuclear lamins: thin filaments with major functions. Trends Cell Biol 28(1):34–45. https://doi.org/10.1016/j.tcb.2017.08.004
Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD (2008) Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 22(7):832–853. https://doi.org/10.1101/gad.1652708
Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD (2010a) Nuclear lamins. Cold Spring Harb Perspect Biol 2(11):a000547. https://doi.org/10.1101/cshperspect.a000547
Dechat T, Gesson K, Foisner R (2010b) Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harb Symp Quant Biol 75:533–543. https://doi.org/10.1101/sqb.2010.75.018
Dittmer TA, Misteli T (2011) The lamin protein family. Genome Biol 12(5):222. https://doi.org/10.1186/gb-2011-12-5-222
Donnaloja F, Jacchetti E, Soncini M, Raimondi MT (2019) Mechanosensing at the nuclear envelope by nuclear pore complex stretch activation and its effect in physiology and pathology. Front Physiol 10:896. https://doi.org/10.3389/fphys.2019.00896
Du J, Fan Y, Guo Z, Wang Y, Zheng X, Huang C, Liang B, Gao L, Cao Y, Chen Y, Zhang X, Li L, Xu L, Wu C, Weitz DA, Feng X (2019) Compression generated by a 3D supracellular actomyosin cortex promotes embryonic stem cell colony growth and expression of Nanog and Oct4. Cell Syst 9(2):214–220.e215. https://doi.org/10.1016/j.cels.2019.05.008
Dupont S, Wickstrom SA (2022) Mechanical regulation of chromatin and transcription. Nat Rev Genet 23(10):624–643. https://doi.org/10.1038/s41576-022-00493-6
Eckersley-Maslin MA, Bergmann JH, Lazar Z, Spector DL (2013) Lamin A/C is expressed in pluripotent mouse embryonic stem cells. Nucleus 4(1):53–60. https://doi.org/10.4161/nucl.23384
Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, Shanahan CM, Trepat X, Navajas D, Garcia-Manyes S, Roca-Cusachs P (2017) Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171(6):1397–1410e1314. https://doi.org/10.1016/j.cell.2017.10.008
Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126(4):677–689. https://doi.org/10.1016/j.cell.2006.06.044
Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA (2008) Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4(3):e1000039. https://doi.org/10.1371/journal.pgen.1000039
Fletcher DA, Mullins RD (2010) Cell mechanics and the cytoskeleton. Nature 463(7280):485–492. https://doi.org/10.1038/nature08908
Galie PA, Georges PC, Janmey PA (2022) How do cells stiffen? Biochem J 479(17):1825–1842. https://doi.org/10.1042/BCJ20210806
Gan Z, Ding L, Burckhardt CJ, Lowery J, Zaritsky A, Sitterley K, Mota A, Costigliola N, Starker CG, Voytas DF, Tytell J, Goldman RD, Danuser G (2016) Vimentin intermediate filaments template microtubule networks to enhance persistence in cell polarity and directed migration. Cell Syst 3(3):252–263.e258. https://doi.org/10.1016/j.cels.2016.08.007
Garcia DA, Fettweis G, Presman DM, Paakinaho V, Jarzynski C, Upadhyaya A, Hager GL (2021) Power-law behavior of transcription factor dynamics at the single-molecule level implies a continuum affinity model. Nucleic Acids Res 49(12):6605–6620. https://doi.org/10.1093/nar/gkab072
Gittes F, Mickey B, Nettleton J, Howard J (1993) Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol 120(4):923–934. https://doi.org/10.1083/jcb.120.4.923
Goldstein I, Hager GL (2018) Dynamic enhancer function in the chromatin context. Wiley Interdiscip Rev Syst Biol Med 10(1). https://doi.org/10.1002/wsbm.1390
Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453(7197):948–951. https://doi.org/10.1038/nature06947
Hamouda MS, Labouesse C, Chalut KJ (2020) Nuclear mechanotransduction in stem cells. Curr Opin Cell Biol 64:97–104. https://doi.org/10.1016/j.ceb.2020.05.005
Heidemann SR, Wirtz D (2004) Towards a regional approach to cell mechanics. Trends Cell Biol 14(4):161–166. https://doi.org/10.1016/j.tcb.2004.02.003
Heo SJ, Cosgrove BD, Dai EN, Mauck RL (2018) Mechano-adaptation of the stem cell nucleus. Nucleus 9(1):9–19. https://doi.org/10.1080/19491034.2017.1371398
Hoffman LM, Smith MA, Jensen CC, Yoshigi M, Blankman E, Ullman KS, Beckerle MC (2020) Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol Biol Cell 31(16):1774–1787. https://doi.org/10.1091/mbc.E19-01-0027
Hohmann T, Dehghani F (2019) The cytoskeleton-A complex interacting meshwork. Cells 8(4):362. https://doi.org/10.3390/cells8040362
Howard J (2001) Mechanics of motor proteins and the cytoskeleton. Sinauer Associates, Sunderland, MA
Hsia CR, McAllister J, Hasan O, Judd J, Lee S, Agrawal R, Chang CY, Soloway P, Lammerding J (2022) Confined migration induces heterochromatin formation and alters chromatin accessibility. iScience 25(9):104978. https://doi.org/10.1016/j.isci.2022.104978
Hu S, Chen J, Butler JP, Wang N (2005) Prestress mediates force propagation into the nucleus. Biochem Biophys Res Commun 329(2):423–428. https://doi.org/10.1016/j.bbrc.2005.02.026
Infante E, Etienne-Manneville S (2022) Intermediate filaments: integration of cell mechanical properties during migration. Front Cell Dev Biol 10:951816. https://doi.org/10.3389/fcell.2022.951816
Irgen-Gioro S, Yoshida S, Walling V, Chong S (2022) Fixation can change the appearance of phase separation in living cells. Elife 11:e79903. https://doi.org/10.7554/eLife.79903
Jacobson EC, Perry JK, Long DS, Olins AL, Olins DE, Wright BE, Vickers MH, O'Sullivan JM (2018) Migration through a small pore disrupts inactive chromatin organization in neutrophil-like cells. BMC Biol 16(1):142. https://doi.org/10.1186/s12915-018-0608-2
Jahed Z, Soheilypour M, Peyro M, Mofrad MR (2016) The LINC and NPC relationship - it's complicated! J Cell Sci 129(17):3219–3229. https://doi.org/10.1242/jcs.184184
Kalukula Y, Stephens AD, Lammerding J, Gabriele S (2022) Mechanics and functional consequences of nuclear deformations. Nat Rev Mol Cell Biol 23(9):583–602. https://doi.org/10.1038/s41580-022-00480-z
Keeling MC, Flores LR, Dodhy AH, Murray ER, Gavara N (2017) Actomyosin and vimentin cytoskeletal networks regulate nuclear shape, mechanics and chromatin organization. Sci Rep 7(1):5219. https://doi.org/10.1038/s41598-017-05467-x
Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y (2011) Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science 334(6063):1706–1710. https://doi.org/10.1126/science.1211222
Kim Y, Zheng X, Zheng Y (2013) Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res 23(12):1420–1423. https://doi.org/10.1038/cr.2013.118
Kindberg A, Hu JK, Bush JO (2020) Forced to communicate: integration of mechanical and biochemical signaling in morphogenesis. Curr Opin Cell Biol 66:59–68. https://doi.org/10.1016/j.ceb.2020.05.004
Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT (2006) Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem 281(35):25768–25780. https://doi.org/10.1074/jbc.M513511200
Leduc C, Etienne-Manneville S (2017) Regulation of microtubuleassociated motors drives intermediate filament network polarization. J Cell Biol 216(6):1689–1703. https://doi.org/10.1083/jcb.201607045
Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D (2007) Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J 93(7):2542–2552. https://doi.org/10.1529/biophysj.106.102426
Li Y, Almassalha LM, Chandler JE, Zhou X, Stypula-Cyrus YE, Hujsak KA, Roth EW, Bleher R, Subramanian H, Szleifer I, Dravid VP, Backman V (2017) The effects of chemical fixation on the cellular nanostructure. Exp Cell Res 358(2):253–259. https://doi.org/10.1016/j.yexcr.2017.06.022
Lionnet T, Wu C (2021) Single-molecule tracking of transcription protein dynamics in living cells: seeing is believing, but what are we seeing? Curr Opin Genet Dev 67:94–102. https://doi.org/10.1016/j.gde.2020.12.001
Lochs SJA, Kefalopoulou S, Kind J (2019) Lamina associated domains and gene regulation in development and cancer. Cells 8(3):271. https://doi.org/10.3390/cells8030271
Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW et al (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38(4):431–440. https://doi.org/10.1038/ng1760
Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J (2011) The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem 286(30):26743–26753. https://doi.org/10.1074/jbc.M111.233700
Malashicheva A, Perepelina K (2021) Diversity of nuclear lamin A/C Action as a key to tissue-specific regulation of cellular identity in health and disease. Front Cell Dev Biol 9:761469. https://doi.org/10.3389/fcell.2021.761469
Matsuda A, Mofrad MRK (2022) On the nuclear pore complex and its emerging role in cellular mechanotransduction. APL Bioeng 6(1):011504. https://doi.org/10.1063/5.0080480
Mazza D, Stasevich TJ, Karpova TS, McNally JG (2012) Monitoring dynamic binding of chromatin proteins in vivo by fluorescence correlation spectroscopy and temporal image correlation spectroscopy. Methods Mol Biol 833:177–200. https://doi.org/10.1007/978-1-61779-477-3_12
Mikuni S, Tamura M, Kinjo M (2007) Analysis of intranuclear binding process of glucocorticoid receptor using fluorescence correlation spectroscopy. FEBS Lett 581(3):389–393. https://doi.org/10.1016/j.febslet.2006.12.038
Mizuno D, Tardin C, Schmidt CF, Mackintosh FC (2007) Nonequilibrium mechanics of active cytoskeletal networks. Science 315(5810):370–373. https://doi.org/10.1126/science.1134404
Morisaki T, Müller WG, Golob N, Mazza D, McNally JG (2014) Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat Commun 5(1):4456. https://doi.org/10.1038/ncomms5456
Muehlich S, Hermanns C, Meier MA, Kircher P, Gudermann T (2016) Unravelling a new mechanism linking actin polymerization and gene transcription. Nucleus 7(2):121–125. https://doi.org/10.1080/19491034.2016.1171433
Mueller F, Mazza D, Stasevich TJ, McNally JG (2010) FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr Opin Cell Biol 22(3):403–411. https://doi.org/10.1016/j.ceb.2010.03.002
Murphy WL, McDevitt TC, Engler AJ (2014) Materials as stem cell regulators. Nat Mater 13(6):547–557. https://doi.org/10.1038/nmat3937
Nava MM, Miroshnikova YA, Biggs LC, Whitefield DB, Metge F, Boucas J, Vihinen H, Jokitalo E, Li X, Garcia Arcos JM, Hoffmann B, Merkel R, Niessen CM, Dahl KN, Wickstrom SA (2020) Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181(4):800–817.e822. https://doi.org/10.1016/j.cell.2020.03.052
Nmezi B, Xu J, Fu R, Armiger TJ, Rodriguez-Bey G, Powell JS, Ma H, Sullivan M, Tu Y, Chen NY, Young SG, Stolz DB, Dahl KN, Liu Y, Padiath QS (2019) Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina. Proc Natl Acad Sci U S A 116(10):4307–4315. https://doi.org/10.1073/pnas.1810070116
Paakinaho V, Presman DM, Ball DA, Johnson TA, Schiltz RL, Levitt P, Mazza D, Morisaki T, Karpova TS, Hager GL (2017) Singlemolecule analysis of steroid receptor and cofactor action in living cells. Nat Commun 8:15896. https://doi.org/10.1038/ncomms15896
Pallavicini C, Levi V, Wetzler DE, Angiolini JF, Bensenor L, Desposito MA, Bruno L (2014) Lateral motion and bending of microtubules studied with a new single-filament tracking routine in living cells. Biophys J 106(12):2625–2635. https://doi.org/10.1016/j.bpj.2014.04.046
Pascual-Reguant L, Blanco E, Galan S, Le Dily F, Cuartero Y, Serra-Bardenys G, Di Carlo V, Iturbide A, Cebria-Costa JP, Nonell L, de Herreros AG, Di Croce L, Marti-Renom MA, Peiro S (2018) Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat Commun 9(1):3420. https://doi.org/10.1038/s41467-018-05912-z
Patange S, Ball DA, Wan Y, Karpova TS, Girvan M, Levens D, Larson DR (2022) MYC amplifies gene expression through global changes in transcription factor dynamics. Cell Rep 38(4):110292. https://doi.org/10.1016/j.celrep.2021.110292
Patteson AE, Vahabikashi A, Goldman RD, Janmey PA (2020) Mechanical and non-mechanical functions of filamentous and non-filamentous vimentin. BioEssays 42(11):e2000078. https://doi.org/10.1002/bies.202000078
Pegoraro AF, Janmey P, Weitz DA (2017) Mechanical properties of the cytoskeleton and cells. Cold Spring Harb Perspect Biol 9(11):a022038. https://doi.org/10.1101/cshperspect.a022038
Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M, Wessels L, van Steensel B (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38(4):603–613. https://doi.org/10.1016/j.molcel.2010.03.016
Petzold J, Gentleman E (2021) Intrinsic mechanical cues and their impact on stem cells and embryogenesis. Front Cell Dev Biol 9:761871. https://doi.org/10.3389/fcell.2021.761871
Pimm ML, Henty-Ridilla JL (2021) New twists in actin-microtubule interactions. Mol Biol Cell 32(3):211–217. https://doi.org/10.1091/mbc.E19-09-0491
Poh YC, Shevtsov SP, Chowdhury F, Wu DC, Na S, Dundr M, Wang N (2012) Dynamic force-induced direct dissociation of protein complexes in a nuclear body in living cells. Nat Commun 3:866. https://doi.org/10.1038/ncomms1873
Presman DM, Ball DA, Paakinaho V, Grimm JB, Lavis LD, Karpova TS, Hager GL (2017) Quantifying transcription factor binding dynamics at the single-molecule level in live cells. Methods 123:76–88. https://doi.org/10.1016/j.ymeth.2017.03.014
Ptak C, Aitchison JD, Wozniak RW (2014) The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol 28:46–53. https://doi.org/10.1016/j.ceb.2014.02.001
Rashid F, Liu W, Wang Q, Ji B, Irudayaraj J, Wang N (2023) Mechanomemory in protein diffusivity of chromatin and nucleoplasm after force cessation. Proc Natl Acad Sci U S A 120(13):e2221432120. https://doi.org/10.1073/pnas.2221432120
Reddy KL, Zullo JM, Bertolino E, Singh H (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452(7184):243–247. https://doi.org/10.1038/nature06727
Romero JJ, De Rossi MC, Oses C, Echegaray CV, Verneri P, Francia M, Guberman A, Levi V (2022) Nucleus-cytoskeleton communication impacts on OCT4-chromatin interactions in embryonic stem cells. BMC Biol 20(1):6. https://doi.org/10.1186/s12915-021-01207-w
Sapra KT, Medalia O (2021) Bend, push, stretch: remarkable structure and mechanics of single intermediate filaments and meshworks. Cells 10(8):1960. https://doi.org/10.3390/cells10081960
Schmidt HB, Gorlich D (2016) Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci 41(1):46–61. https://doi.org/10.1016/j.tibs.2015.11.001
Schmiedeberg L, Weisshart K, Diekmann S, Meyer zuHoerste G, Hemmerich P (2004) High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell 15(6):2819-2833. https://doi.org/10.1091/mbc.e03-11-0827
Schuller AP, Wojtynek M, Mankus D, Tatli M, Kronenberg-Tenga R, Regmi SG, Dip PV, Lytton-Jean AKR, Brignole EJ, Dasso M, Weis K, Medalia O, Schwartz TU (2021) The cellular environment shapes the nuclear pore complex architecture. Nature 598(7882):667–671. https://doi.org/10.1038/s41586-021-03985-3
See K, Lan Y, Rhoades J, Jain R, Smith CL, Epstein JA (2019) Lineage-specific reorganization of nuclear peripheral heterochromatin and H3K9me2 domains. Development 146(3):dev174078. https://doi.org/10.1242/dev.174078
Shelden E, Wadsworth P (1993) Observation and quantification of individual microtubule behavior in vivo: microtubule dynamics are cell-type specific. J Cell Biol 120(4):935–945. https://doi.org/10.1083/jcb.120.4.935
Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD (2008) The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev 22(24):3409–3421. https://doi.org/10.1101/gad.1735208
Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD (2015) Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell 26(22):4075–4086. https://doi.org/10.1091/mbc.E15-07-0461
Smith MA, Blankman E, Jensen CC, Hoffman LM, Ullman KS, Beckerle MC (2022) Nuclear pore complexes concentrate on Actin/ LINC/Lamin nuclear lines in response to mechanical stress in a SUN1 dependent manner. Heliyon 8(12):e12147. https://doi.org/10.1016/j.heliyon.2022.e12147
Smoler M, Coceano G, Testa I, Bruno L, Levi V (2020) Apparent stiffness of vimentin intermediate filaments in living cells and its relation with other cytoskeletal polymers. Biochim Biophys Acta Mol Cell Res 1867(8):118726. https://doi.org/10.1016/j.bbamcr.2020.118726
Song Y, Soto J, Chen B, Hoffman T, Zhao W, Zhu N, Peng Q, Liu L, Ly C, Wong PK, Wang Y, Rowat AC, Kurdistani SK, Li S (2022) Transient nuclear deformation primes epigenetic state and promotes cell reprogramming. Nat Mater 21(10):1191–1199. https://doi.org/10.1038/s41563-022-01312-3
Srivastava LK, Ju Z, Ghagre A, Ehrlicher AJ (2021) Spatial distribution of lamin A/C determines nuclear stiffness and stressmediated deformation. J Cell Sci 134(10):jcs248559. https://doi.org/10.1242/jcs.248559
Starr DA, Fridolfsson HN (2010) Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26:421–444. https://doi.org/10.1146/annurev-cellbio-100109-104037
Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF (2017) Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell 28(14):1984–1996. https://doi.org/10.1091/mbc.e16-09-0653
Stortz M, Presman DM, Bruno L, Annibale P, Dansey MV, Burton G, Gratton E, Pecci A, Levi V (2017) Mapping the dynamics of the glucocorticoid receptor within the nuclear landscape. Sci Rep 7(1):6219. https://doi.org/10.1038/s41598-017-06676-0
Stortz M, Angiolini J, Mocskos E, Wolosiuk A, Pecci A, Levi V (2018) Mapping the dynamical organization of the cell nucleus through fluorescence correlation spectroscopy. Methods 140−141:10–20. https://doi.org/10.1016/j.ymeth.2017.12.008
Stortz M, Oses C, Vazquez Echegaray C, Pecci A, Guberman A, Presman DM, Levi V (2022) SOX2 modulates the nuclear organization and transcriptional activity of the glucocorticoid receptor. J Mol Biol 434(24):167869. https://doi.org/10.1016/j.jmb.2022.167869
Sun J, Chen J, Mohagheghian E, Wang N (2020) Force-induced gene up-regulation does not follow the weak power law but depends on H3K9 demethylation. Sci Adv 6(14):eaay9095. https://doi.org/10.1126/sciadv.aay9095
Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, Rehfeldt F, Speicher DW, Discher DE (2013) Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341(6149):1240104. https://doi.org/10.1126/science.1240104
Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N (2016) Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 15(12):1287–1296. https://doi.org/10.1038/nmat4729
Texari L, Stutz F (2015) Sumoylation and transcription regulation at nuclear pores. Chromosoma 124(1):45–56. https://doi.org/10.1007/s00412-014-0481-x
Totaro A, Panciera T, Piccolo S (2018) YAP/TAZ upstream signals and downstream responses. Nat Cell Biol 20(8):888–899. https://doi.org/10.1038/s41556-018-0142-z
Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O (2017) The molecular architecture of lamins in somatic cells. Nature 543(7644):261–264. https://doi.org/10.1038/nature21382
Vahabikashi A, Adam SA, Medalia O, Goldman RD (2022) Nuclear lamins: structure and function in mechanobiology. APL Bioeng 6(1):011503. https://doi.org/10.1063/5.0082656
van Mameren J, Vermeulen KC, Gittes F, Schmidt CF (2009) Leveraging single protein polymers to measure flexural rigidity. J Phys Chem B 113(12):3837–3844. https://doi.org/10.1021/jp808328a
Verneri P, Vazquez Echegaray C, Oses C, Stortz M, Guberman A, Levi V (2020) Dynamical reorganization of the pluripotency transcription factors Oct4 and Sox2 during early differentiation of embryonic stem cells. Sci Rep 10(1):5195. https://doi.org/10.1038/s41598-020-62235-0
Versaevel M, Braquenier JB, Riaz M, Grevesse T, Lantoine J, Gabriele S (2014) Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci Rep 4:7362. https://doi.org/10.1038/srep07362
Vining KH, Mooney DJ (2017) Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol 18(12):728–742. https://doi.org/10.1038/nrm.2017.108
Wang N, Tytell JD, Ingber DE (2009) Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 10(1):75–82. https://doi.org/10.1038/nrm2594
Wang Y, Elsherbiny A, Kessler L, Cordero J, Shi H, Serke H, Lityagina O, Trogisch FA, Mohammadi MM, El-Battrawy I, Backs J, Wieland T, Heineke J, Dobreva G (2022) Lamin A/C-dependent chromatin architecture safeguards naive pluripotency to prevent aberrant cardiovascular cell fate and function. Nat Commun 13(1):6663. https://doi.org/10.1038/s41467-022-34366-7
White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, Mocskos E, Bruno L, Bissiere S, Levi V, Plachta N (2016) Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165(1):75–87. https://doi.org/10.1016/j.cell.2016.02.032
Wong X, Stewart CL (2020) The laminopathies and the insights they provide into the structural and functional organization of the nucleus. Annu Rev Genomics Hum Genet 21:263–288. https://doi.org/10.1146/annurev-genom-121219-083616
Wong X, Hoskins VE, Melendez-Perez AJ, Harr JC, Gordon M, Reddy KL (2021) Lamin C is required to establish genome organization after mitosis. Genome Biol 22(1):305. https://doi.org/10.1186/s13059-021-02516-7
Wu J, Lewis AH, Grandl J (2017) Touch, Tension, and Transduction - The Function and Regulation of Piezo Ion Channels. Trends Biochem Sci 42(1):57–71. https://doi.org/10.1016/j.tibs.2016.09.004
Wu PH, Aroush DR, Asnacios A, Chen WC, Dokukin ME, Doss BL, Durand-Smet P, Ekpenyong A, Guck J, Guz NV, Janmey PA, Lee JSH, Moore NM, Ott A, Poh YC, Ros R, Sander M, Sokolov I, Staunton JR et al (2018) A comparison of methods to assess cell mechanical properties. Nat Methods 15(7):491–498. https://doi.org/10.1038/s41592-018-0015-1
Wu H, Shen Y, Sivagurunathan S, Weber MS, Adam SA, Shin JH, Fredberg JJ, Medalia O, Goldman R, Weitz DA (2022) Vimentin intermediate filaments and filamentous actin form unexpected interpenetrating networks that redefine the cell cortex. Proc Natl Acad Sci U S A 119(10):e2115217119. https://doi.org/10.1073/pnas.2115217119
Xia S, Lim YB, Zhang Z, Wang Y, Zhang S, Lim CT, Yim EKF, Kanchanawong P (2019) Nanoscale architecture of the cortical actin cytoskeleton in embryonic stem cells. Cell Rep 28(5):1251–1267 e1257. https://doi.org/10.1016/j.celrep.2019.06.089
Yanagida T, Nakase M, Nishiyama K, Oosawa F (1984) Direct observation of motion of single F-actin filaments in the presence of myosin. Nature 307(5946):58–60. https://doi.org/10.1038/307058a0
Yoon M, Moir RD, Prahlad V, Goldman RD (1998) Motile properties of vimentin intermediate filament networks in living cells. J Cell Biol 143(1):147–157. https://doi.org/10.1083/jcb.143.1.147
Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, Halavatyi A, Hummer G, Mahamid J, Kosinski J, Beck M (2021) Nuclear pores dilate and constrict in cellulo. Science 374(6573):eabd9776. https://doi.org/10.1126/science.abd9776
Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J (2013) Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum Mol Genet 22(12):2335–2349. https://doi.org/10.1093/hmg/ddt079
Acknowledgements
This research was supported by ANPCyT (PICT 2020-00818, and PICT-2018-1921 to V.L.) and Universidad de Buenos Aires (UBACyT 20020190100101BA to V.L.). We acknowledge Juan J Romero for the 3D images of ESCs and Diego Presman for his valuable comments. Due to space constraints, we could not discuss many interesting works in the area; we apologize to their authors.
Author information
Authors and Affiliations
Contributions
The first draft of the manuscript was written by [Valeria Levi]. [Camila Oses], [María Cecilia De Rossi]. [Luciana Bruno] and [María Candelaria Diaz] prepared the figures. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Oses, C., De Rossi, M.C., Bruno, L. et al. From the membrane to the nucleus: mechanical signals and transcription regulation. Biophys Rev 15, 671–683 (2023). https://doi.org/10.1007/s12551-023-01103-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-023-01103-3