Abstract
Introduction
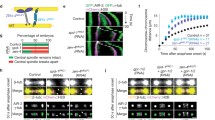
The microtubule motor protein kinesin-5 is well known to establish the bipolar spindle by outward sliding of antiparallel interpolar microtubules. In yeast, kinesin-5 also facilitates chromosome alignment “congression” at the spindle equator by preferentially depolymerizing long kinetochore microtubules (kMTs). The motor protein kinesin-8 has also been linked to chromosome congression. Therefore, we sought to determine whether kinesin-5 or kinesin-8 facilitates chromosome congression in insect spindles.
Methods
RNAi of the kinesin-5 Klp61F and kinesin-8 Klp67A were performed separately in Drosophila melanogaster S2 cells to test for inhibited chromosome congression. Klp61F RNAi, Klp67A RNAi, and control metaphase mitotic spindles expressing fluorescent tubulin and fluorescent Cid were imaged, and their fluorescence distributions were compared.
Results
RNAi of Klp61F with a weak Klp61F knockdown resulted in longer kMTs and less congressed kinetochores compared to control over a range of conditions, consistent with kinesin-5 length-dependent depolymerase activity. RNAi of the kinesin-8 Klp67A revealed that kMTs relative to the spindle lengths were not longer compared to control, but rather that the spindles were longer, indicating that Klp67A acts preferentially as a length-dependent depolymerase on interpolar microtubules without significantly affecting kMT length and chromosome congression.
Conclusions
This study demonstrates that in addition to establishing the bipolar spindle, kinesin-5 regulates kMT length to facilitate chromosome congression in insect spindles. It expands on previous yeast studies, and it expands the role of kinesin-5 to include kMT assembly regulation in eukaryotic mitosis.



Similar content being viewed by others
Abbreviations
- kMT:
-
Kinetochore microtubule
- iMT:
-
Interpolar microtubule
- RNAi:
-
RNA interference
- dsRNA:
-
Double stranded RNA
- NEB:
-
Nuclear envelope breakdown
- AO:
-
Anaphase onset
- PEF:
-
Polar ejection force
References
Ault, J. G., and C. L. Rieder. Centrosome and kinetochore movement during mitosis. Curr. Opin. Cell Biol. 6:41–49, 1994.
Brinkley, B. R., R. P. Zinkowski, W. L. Mollon, F. M. Davis, M. A. Pisegna, M. Pershouse, and P. N. Rao. Movement and segregation of kinetochores experimentally detached from mammalian chromosomes. Nature 336:251–254, 1988.
Brouhard, G. J., and A. J. Hunt. Microtubule movements on the arms of mitotic chromosomes: polar ejection forces quantified in vitro. Proc. Natl. Acad. Sci. USA 102:13903–139038, 2005.
Brust-Mascher, I., G. Civelekoglu-Scholey, M. Kwon, A. Mogilner, and J. M. Scholey. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA 101:15938–15943, 2004.
Brust-Mascher, I., P. Sommi, D. K. Cheerambathur, and J. M. Scholey. Kinesin-5 – dependent Poleward Flux and Spindle Length Control in Drosophila Embryo Mitosis. Mol. Biol. Cell 20:1749–1762, 2009.
Cassimeris, L., C. L. Rieder, and E. D. Salmon. Microtubule assembly and kinetochore directional instability in vertebrate monopolar spindles: implications for the mechanism of chromosome congression. J. Cell Sci. 107:285–297, 1994.
Chacón, J. M., S. Mukherjee, B. M. Schuster, D. J. Clarke, and M. K. Gardner. Pericentromere tension is self-regulated by spindle structure in metaphase. J. Cell Biol. 205:313–324, 2014.
Chen, Y., and W. O. Hancock. Kinesin-5 is a microtubule polymerase. Nat. Commun. Nature Publishing Group 6:1–10, 2015.
Clemens, J.C. et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci., USA 97:6499–503, 2000.
Demchouk, A. O., M. K. Gardner, and D. J. Odde. Microtubule tip tracking and tip structures at the nanometer scale using digital fluorescence microscopy. Cell. Mol. Bioeng. 4:192–204, 2011.
Ferenz, N. P., A. Gable, and P. Wadsworth. Mitotic functions of kinesin-5. Semin. Cell Dev. Biol. Elsevier Ltd 21:255–259, 2010.
Fridman, V., A. Gerson-Gurwitz, O. Shapira, N. Movshovich, S. Lakämper, C. Schmidt, and L. Gheber. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J. Cell Sci. 126:4147–4159, 2013.
Funabiki, H., and A. W. Murray. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102:411–424, 2000.
Gardner, M. K., C. G. Pearson, B. L. Sprague, T. R. Zarzar, K. Bloom, E. D. Salmon, and D. J. Odde. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol. Biol. Cell 16:3764–3775, 2005.
Gardner, M.K., D.C. Bouck, L.V. Paliulis, J.B. Meehl, E.T. O’Toole, J. Haase, A. Soubry, A.P. Joglekar, M. Windey, E.D. Salmon, K. Bloom, and D. J. Odde. Chromosome congression by kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell 135:894–906, 2008.
Gerson-Gurwitz, A., C. Thiede, N. Movshovich, V. Fridman, M. Podolskaya, T. Danieli, S. Lakämper, D. R. Klopfenstein, C. F. Schmidt, and L. Gheber. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 30:4942–4954, 2011.
Goodwin, S. S., and R. D. Vale. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143:263–274, 2010.
Goshima, G., et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316:417–421, 2007.
Goshima, G., and R. D. Vale. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016, 2003.
Goshima, G., R. Wollman, N. Stuurman, J. M. Scholey, and R. D. Vale. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988, 2005.
Heck, M., A. Pereira, P. Pesavento, Y. Yannoni, A. C. Spradling, and L. S. Goldstein. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J. Cell Biol. 123:665–679, 1993.
Henikoff, S., K. Ahmad, J.S. Platero, and B. van Steensel. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci., USA 97:716–721, 2000.
Hoyt, M. A., L. He, K. K. Loo, and W. S. Saunders. Kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 118:109–120, 1992.
Inoué, S., and E. D. Salmon. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 6:1619–1640, 1995.
Ke, K., J. Cheng, and A. J. Hunt. The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr. Biol. Elsevier Ltd 19:807–815, 2009.
Levesque, A. A., and D. A. Compton. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell. Biol. 154:1135–1146, 2001.
Maiato, H., P. J. Hergert, S. Moutinho-Periera, Y. Dong, K. J. Vandenbeldt, C. L. Rieder, and B. F. McEwen. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma 115:469–480, 2006.
Maiato, H., C. E. Sunkel, and W. C. Earnshaw. Dissecting mitosis by RNAi in Drosophila tissue culture cells. Biol. Proced. Online 5:153–161, 2003.
Mayr, M. I., S. Hümmer, J. Bormann, T. Grüner, S. Adio, G. Woehlke, and T. U. Mayer. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 17:488–498, 2007.
McCoy, K. M., E. S. Tubman, A. Claas, D. Tank, S. A. Clancy, E. T. O’Toole, J. Berman, and D. J. Odde. Physical limits on kinesin-5 mediated chromosome congression in the smallest mitotic spindles. Mol. Biol. Cell 26:3999–4014, 2015.
Mische, S., Y. He, L. Ma, M. Li, M. Serr, and T. S. Hays. Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol. Biol. Cell 19:4918–4929, 2008.
Moore, D. S., and G. McCabe. Introduction to the practice of statistics, Vol. Sixth. New York: WH Freeman and Company, 2009.
O’Connell, C. B., J. Loncarek, P. Hergert, A. Kourtidis, D. S. Conklin, and A. Khodjakov. The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes. J. Cell Biol. 183:29–36, 2008.
O’Connell, C. B., J. Lončarek, P. Kaláb, and A. Khodjakov. Relative contributions of chromatin and kinetochores to mitotic spindle assembly. J. Cell Biol. 187:43–51, 2009.
Orth, J. D., Y. Tang, J. Shi, C. T. Loy, C. Amendt, C. Wilm, F. T. Zenke, and T. J. Mitchison. Quantitative live imaging of cancer and normal cells treated with Kinesin-5 inhibitors indicates significant differences in phenotypic responses and cell fate. Mol. Cancer Ther. 7:3480–3489, 2008.
Rieder, C. L., E. A. Davison, L. C. Jensen, L. Cassimeris, and E. D. Salmon. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103:581–591, 1986.
Rieder, C. L., and H. Maiato. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7:637–651, 2004.
Rogers, S. L., G. C. Rogers, D. J. Sharp, and R. D. Vale. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884, 2002.
Roof, D. M., P. B. Meluh, and M. D. Rose. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 118:95–108, 1992.
Roostalu, J., C. Hentrich, P. Bieling, I. A. Telley, E. Schiebel, and T. Surrey. Directional switching of the kinesin Cin8 through motor coupling. Science 332:94–99, 2011.
Savoian, M. S., M. K. Gatt, M. G. Riparbelli, G. Callaini, and D. M. Glover. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J. Cell Sci. 117:3669–3677, 2004.
Savoian, M. S., and D. M. Glover. Drosophila Klp67A binds prophase kinetochores to subsequently regulate congression and spindle length. J. Cell Sci. 123:767–776, 2010.
Sawin, K., K. LeGuellec, M. Phillipe, and T. J. Mitchison. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359:540–543, 1992.
Seetapun, D., B. T. Castle, A. J. McIntyre, P. T. Tran, and D. J. Odde. Estimating the microtubule GTP cap size in vivo. Curr. Biol. 22:1681–1687, 2012.
Sharp, D.J., K.L. McDonald, H.M. Brown, H.J. Matthies, C. Walczak, R.D. Vale, T.J. Mitchison, and J.M. Scholey. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144:125–138, 1999
Sharp, D. J., K. R. Yu, J. C. Sisson, W. Sullivan, and J. M. Scholey. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1:51–54, 1999.
Sprague, B. L., C. G. Pearson, P. S. Maddox, K. S. Bloom, E. D. Salmon, and D. J. Odde. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys. J. Elsevier 84:3529–3546, 2003.
Straight, A. F., J. W. Sedat, and A. W. Murray. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143:687–694, 1998.
Stumpff, J., G. von Dassow, M. Wagenbach, C. Asbury, and L. Wordeman. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell 14:252–262, 2008.
Stumpff, J., M. Wagenbach, A. Franck, C.L. Asbury, and L. Wordeman. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell Elsevier Inc. 22:1017–29, 2012.
Vale, R. D., J. A. Spudich, and E. R. Griffis. Dynamics of myosin, microtubules, and Kinesin-6 at the cortex during cytokinesis in Drosophila S2 cells. J. Cell Biol. 186:727–738, 2009.
Varga, V., J. Helenius, K. Tanaka, A. A. Hyman, T. U. Tanaka, and J. Howard. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8:957–962, 2006.
Wargacki, M. M., J. C. Tay, E. G. Muller, C. L. Asbury, and T. N. Davis. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle 9:2581–2588, 2010.
Winey, M., C. L. Mamay, E. T. O’Toole, D. N. Mastronarde, T. H. Giddings, Jr, K. L. McDonald, and J. R. McIntosh. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615, 1995.
Wise, D. A., and B. R. Brinkley. Mitosis in cells with unreplicated genomes (MUGs): spindle assembly and behavior of centromere fragments. Cell Motil. Cytoskeleton 36:291–302, 1997.
Acknowledgments
We thank Professor Lawrence Goldstein for providing us with rat-anti-Klp61F antibody. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award No. R01GM071522 and R01GM076177 to D.J.O. and Award RO1GM044757 to T.S.H. E.T. was a recipient of a University of Minnesota Interdisciplinary Doctoral Fellowship through the Institute for Advanced Study.
Author Contributions
E.T. conducted RNAi experiments, collected images, wrote analysis algorithms, ran statistical tests, analyzed and interpreted results, prepared figures, and wrote paper. E.T. and Y.H. designed primers, prepared dsRNA, and ran Western Blot. Y.H. contributed to intellectual ideas. T.H. and D.O., co-principal investigators, oversaw the project and contributed to intellectual ideas.
Conflicts of interest
Emily Tubman, Yungui He, Thomas S. Hays, and David J. Odde declare that they have no conflicts of interest.
Ethical Standards
No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor William O. Hancock oversaw the review of this article.
Thomas S. Hays and David J. Odde are co-senior authors.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tubman, E., He, Y., Hays, T.S. et al. Kinesin-5 Mediated Chromosome Congression in Insect Spindles. Cel. Mol. Bioeng. 11, 25–36 (2018). https://doi.org/10.1007/s12195-017-0500-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-017-0500-0