Abstract
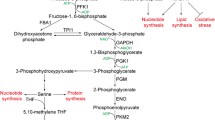
To survive in the tumour microenvironment, cancer cells undergo rapid metabolic reprograming and adaptability. One of the key characteristics of cancer is increased glycolytic selectivity and decreased oxidative phosphorylation (OXPHOS). Apart from ATP synthesis, glycolysis is also responsible for NADH regeneration and macromolecular biosynthesis, such as amino acid biosynthesis and nucleotide biosynthesis. This allows cancer cells to survive and proliferate even in low-nutrient and oxygen conditions, making glycolytic enzymes a promising target for various anti-cancer agents. Oncogenic activation is also caused by the uncontrolled production and activity of glycolytic enzymes. Nevertheless, in addition to conventional glycolytic processes, some glycolytic enzymes are involved in non-canonical functions such as transcriptional regulation, autophagy, epigenetic changes, inflammation, various signaling cascades, redox regulation, oxidative stress, obesity and fatty acid metabolism, diabetes and neurodegenerative disorders, and hypoxia. The mechanisms underlying the non-canonical glycolytic enzyme activities are still not comprehensive. This review summarizes the current findings on the mechanisms fundamental to the non-glycolytic actions of glycolytic enzymes and their intermediates in maintaining the tumor microenvironment.





Similar content being viewed by others
Abbreviations
- 2-DG:
-
2-deoxy-D-glucose
- 5-TG:
-
5-thioglucose
- 6-PGDH:
-
6-Phosphogluconate dehydrogenase
- 6-PGL:
-
6-Phosphogluconolactonase
- ACC:
-
Acetyl CoA Carboxylase
- ACS 1:
-
Acetyl-CoA synthetase
- AD:
-
Alzheimer’s disease
- AKT S1:
-
AKT1 Substrate 1
- ALS:
-
Amyotrophic lateral sclerosis
- AMF:
-
Autocrine motility factor
- AMFR:
-
Autocrine motility factor receptor
- AMP:
-
Adenosine monophosphate
- AMPK:
-
AMP-activated protein kinase
- ANLS:
-
Astrocyte neuron lactate shuttle
- APC:
-
Axin-Adenomatosis Polyposis Coli
- APP:
-
Amyloid precursor protein
- ARE:
-
Anti-oxidant responsive elements
- ARNT:
-
Aryl hydrocarbon receptor nuclear translocator
- ATM:
-
Ataxia- telangiectasia mutant
- ATP:
-
Adenosine triphosphate
- BECN1:
-
Beclin-1
- c-AMP:
-
Cyclic AMP
- CBP:
-
CREB-binding protein
- CNS:
-
Central nervous system
- CPT 1:
-
Carnitine palmitoyl transferase 1
- Cyt c:
-
-Cytochrome c
- DNA:
-
Deoxyribonucleic acid
- DVL:
-
Dishevelled
- EGF:
-
Epidermal growth factor
- EGFR:
-
Epidermal growth factor receptor
- ENO1:
-
Enolase 1
- ER:
-
Endoplasmic reticulum
- ERK:
-
Extracellular signal-regulated kinase
- ETC:
-
Electron transport chain
- FAS:
-
Fatty acid synthesis
- FDG-PET:
-
Fluorodeoxyglucose positron emission tomography
- FDH:
-
Fumarate dehydrogenase
- FDH:
-
Folate dehydrogenase
- FIH:
-
Factor inhibiting HIF
- FZD:
-
Frizzled
- G-3-P:
-
Glyceraldehyde-3-phosphate
- GAPD:
-
Glyceraldehyde 3-phosphate dehydrogenase
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- G-CSF:
-
Granulocyte colony stimulating factor
- GFAP:
-
Glial fibrillary acidic protein
- GLK:
-
Glucokinase
- GLP 1:
-
Glucagon like peptide 1
- GLUT 1:
-
Glucose transporter 1
- GM CSF:
-
Granulocyte-macrophage colony-stimulating factor
- GPI:
-
Glucose 6 phosphate isomerase
- GR:
-
Glutathione reductase
- Grx:
-
Glutaredoxin
- GSH:
-
Glutathione (Reduced)
- GSSG:
-
Glutathione (Oxidized)
- GSK 3:
-
Glycogen synthase kinase 3
- HDAC:
-
Histone deacetylases
- HGP:
-
Hepatic glucose production
- HIF:
-
Hypoxia-inducible factor
- HRE:
-
Hypoxia-responsive elements
- Hsp:
-
Heat shock protein
- HXK:
-
Hexokinase
- IDH:
-
Isocitrate dehydrogenase
- IGF 1:
-
Insulin-like growth factor 1
- IL:
-
Interleukin
- JNK:
-
Jun N-terminal kinase
- LAP:
-
LC3-associated phagocytosis
- LC 3:
-
Microtubule-associated protein 1 A/1B-light chain 3
- LDH:
-
Lactate dehydrogenase
- LKB 1:
-
Liver kinase B1
- MBP-1:
-
c-Myc binding protein 1
- MCT 1:
-
Monocarboxylate transporter 1
- ME:
-
Malic enzyme
- MIP 2A:
-
Macrophage Inflammatory Protein 2
- MLOC:
-
Mitochondrial lactate oxidation complex
- mTORC:
-
Mammalian target of rapamycin
- NAC:
-
N-Acetyl cystine
- NAD:
-
Nicotinamide adenine dinucleotide
- NADH:
-
Nicotinamide adenine dinucleotide (reduced)
- NAFLD:
-
Non-alcoholic fatty liver disease
- ND:
-
Neurodegenerative disease
- NGF:
-
Nerve Growth Factor
- NOX:
-
NADH oxidase
- NRF-2:
-
Nuclear factor erythroid 2- related factor 2
- OMM:
-
Outer mitochondrial membrane
- OXPHOS:
-
Oxidative phosphorylation
- PC:
-
Prostate cancer
- PCAF:
-
p300/CBP associated Factor
- PD:
-
Perkinson’s disease
- PDC:
-
Pyruvate dehydrogenase complex
- PDK 1:
-
Pyruvate dehydrogenase kinase 1
- PEP:
-
Phosphoenolpyruvate
- PFK:
-
Phosphofructokinase
- PFKL:
-
Phosphofructokinase liver type
- PGK:
-
Phosphoglycerate kinase
- PHD:
-
Prolyl Hydroxylase Domain
- PIKK:
-
Phosphatidylinositol-3- kinase
- PKM 2:
-
Pyruvate kinase M2
- PPP:
-
Pentose phosphate pathway
- PYK 1:
-
Pyruvate Kinase
- RBC:
-
Red blood corpuscles
- RNA:
-
Ribonucleic acid
- ROS:
-
Reactive oxygen species
- SAICAR:
-
Succinyl-5-aminoimidazole-4-carboxamide-1-ribose-5’-phosphate
- SAM:
-
S-Adenosyl methionine
- SCD 1:
-
Stearoyl CoA desaturase 1
- SDH:
-
Succinate dehydrogenase
- SGLT:
-
Sodium glucose co-transporter
- SOD:
-
Superoxide dismutase
- STAT:
-
Signal transducer and activator of transcription
- STZ:
-
Streptozotocin
- T1DM/T2DM:
-
Type 1/2 diabetes mellitus
- TAZ:
-
Yes-associated protein 1
- TCA:
-
Tricarboxylic acid cycle
- THF:
-
Tetrahydrofolic acid
- TFAM:
-
Transcription factor A mitochondrial
- TPA:
-
12-O -tetradecanoyl-phorbol-13- acetate
- Trx:
-
Thioredoxin
- TXNIP:
-
Thioredoxin interacting protein
- ULK 1:
-
Unc-51 like autophagy activating kinase
- UTR:
-
Untranslated region
- VEGF:
-
Vascular endothelial growth factor
- VHL:
-
Von-Hippel-Lindau
- WNT:
-
Wingless-related integration site
- YAP:
-
Yes-associated protein 1
References
Yetkin-Arik, B., Vogels, I. M. C., Nowak-Sliwinska, P., Weiss, A., Houtkooper, R. H., Van Noorden, C. J. F., Klaassen, I., & Schlingemann, R. O. (2019). The role of glycolysis and mitochondrial respiration in the formation and functioning of endothelial tip cells during angiogenesis. Scientific Reports, 9(1), 12608. https://doi.org/10.1038/s41598-019-48676-2.
Lunt, S. Y., & Vander Heiden, M. G. (2011). Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annual Review of Cell and Developmental Biology, 27, 441–464. https://doi.org/10.1146/annurev-cellbio-092910-154237.
Lin, X., Xiao, Z., Chen, T., Liang, S. H., & Guo, H. (2020). Glucose metabolism on tumor plasticity, diagnosis, and treatment. Frontiers in Oncology, 10, 317. https://doi.org/10.3389/fonc.2020.00317.
Canback, B., Andersson, S. G., & Kurland, C. G. (2002). The global phylogeny of glycolytic enzymes. Proceedings of the National Academy of Sciences of the United States of America, 99(9), 6097–6102. https://doi.org/10.1073/pnas.082112499.
Shiratori, R., Furuichi, K., Yamaguchi, M., Miyazaki, N., Aoki, H., Chibana, H., Ito, K., & Aoki, S. (2019). Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner. Scientific Reports, 9(1), 18699. https://doi.org/10.1038/s41598-019-55296-3.
Ganapathy-Kanniappan, S., & Geschwind, J. F. (2013). Tumor glycolysis as a target for cancer therapy: Progress and prospects. Molecular Cancer, 12, 152. https://doi.org/10.1186/1476-4598-12-152.
Poellinger, L., & Johnson, R. S. (2004). HIF-1 and hypoxic response: The plot thickens. Current Opinion in Genetics & Development, 14(1), 81–85. https://doi.org/10.1016/j.gde.2003.12.006.
Schofield, C. J., & Ratcliffe, P. J. (2004). Oxygen sensing by HIF hydroxylases. Nature Reviews Molecular Cell Biology, 5(5), 343–354. https://doi.org/10.1038/nrm1366.
Ohh, M., Park, C. W., Ivan, M., Hoffman, M. A., Kim, T. Y., Huang, L. E., Pavletich, N., Chau, V., & Kaelin, W. G. (2000). Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nature Cell Biology, 2(7), 423–427. https://doi.org/10.1038/35017054.
Semenza, G. L., Jiang, B. H., Leung, S. W., Passantino, R., Concordet, J. P., Maire, P., & Giallongo, A. (1996). Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. The Journal of Biological Chemistry, 271(51), 32529–32537. https://doi.org/10.1074/jbc.271.51.32529.
Ashton, T. M., McKenna, W. G., Kunz-Schughart, L. A., & Higgins, G. S. (2018). Oxidative phosphorylation as an emerging target in cancer therapy. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 24(11), 2482–2490. https://doi.org/10.1158/1078-0432.CCR-17-3070.
Bardella, C., Pollard, P. J., & Tomlinson, I. (2011). SDH mutations in cancer. Biochimica et Biophysica Acta, 1807(11), 1432–1443. https://doi.org/10.1016/j.bbabio.2011.07.003.
Lehtonen, H. J., Kiuru, M., Ylisaukko-Oja, S. K., Salovaara, R., Herva, R., Koivisto, P. A., Vierimaa, O., Aittomäki, K., Pukkala, E., Launonen, V., & Aaltonen, L. A. (2006). Increased risk of cancer in patients with fumarate hydratase germline mutation. Journal of Medical Genetics, 43(6), 523–526. https://doi.org/10.1136/jmg.2005.036400.
Reitman, Z. J., & Yan, H. (2010). Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a crossroads of cellular metabolism. Journal of the National Cancer Institute, 102(13), 932–941. https://doi.org/10.1093/jnci/djq187.
Vander Heiden, M. G., Cantley, L. C., & Thompson, C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science (New York, N.Y.), 324(5930), 1029–1033. https://doi.org/10.1126/science.1160809.
Dang, C. V., & Semenza, G. L. (1999). Oncogenic alterations of metabolism. Trends in Biochemical Sciences, 24(2), 68–72. https://doi.org/10.1016/s0968-0004(98)01344-9.
Muz, B., de la Puente, P., Azab, F., & Azab, A. K. (2015). The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckland, N.Z.), 3, 83–92. https://doi.org/10.2147/HP.S93413.
Selak, M. A., Armour, S. M., MacKenzie, E. D., Boulahbel, H., Watson, D. G., Mansfield, K. D., Pan, Y., Simon, M. C., Thompson, C. B., & Gottlieb, E. (2005). Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell, 7(1), 77–85. https://doi.org/10.1016/j.ccr.2004.11.022.
Moreno, C., Santos, R. M., Burns, R., & Zhang, W. C. (2020). Succinate dehydrogenase and ribonucleic acid networks in cancer and other diseases. Cancers, 12(11), 3237. https://doi.org/10.3390/cancers12113237.
Abou Khouzam, R., Brodaczewska, K., Filipiak, A., Zeinelabdin, N. A., Buart, S., Szczylik, C., Kieda, C., & Chouaib, S. (2021). Tumor hypoxia regulates immune escape/invasion: Influence on angiogenesis and potential impact of hypoxic biomarkers on cancer therapies. Frontiers in Immunology, 11, 613114. https://doi.org/10.3389/fimmu.2020.613114.
Hompland, T., Fjeldbo, C. S., & Lyng, H. (2021). Tumor hypoxia as a barrier in cancer therapy: Why levels matter. Cancers, 13(3), 499. https://doi.org/10.3390/cancers13030499.
Brown, J. M., & Wilson, W. R. (2004). Exploiting tumour hypoxia in cancer treatment. Nature Reviews Cancer, 4(6), 437–447. https://doi.org/10.1038/nrc1367.
Huangyang, P., & Simon, M. C. (2018). Hidden features: Exploring the non-canonical functions of metabolic enzymes. Disease Models & Mechanisms, 11(8), dmm033365. https://doi.org/10.1242/dmm.033365.
Qvit, N., Joshi, A. U., Cunningham, A. D., Ferreira, J. C., & Mochly-Rosen, D. (2016). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein-protein interaction inhibitor reveals a non-catalytic role for GAPDH oligomerization in cell death. The Journal of Biological Chemistry, 291(26), 13608–13621. https://doi.org/10.1074/jbc.M115.711630.
Snaebjornsson, M. T., & Schulze, A. (2018). Non-canonical functions of enzymes facilitate cross-talk between cell metabolic and regulatory pathways. Experimental & Molecular Medicine, 50(4), 1–16. https://doi.org/10.1038/s12276-018-0065-6.
Lavik, A. R., McColl, K. S., Lemos, F. O., Kerkhofs, M., Zhong, F., Harr, M., Schlatzer, D., Hamada, K., Mikoshiba, K., Crea, F., Bultynck, G., Bootman, M. D., Parys, J. B., & Distelhorst, C. W. (2022). A non-canonical role for pyruvate kinase M2 as a functional modulator of Ca2+ signalling through IP3 receptors. Biochimica et Biophysica Acta Molecular Cell Research, 1869(4), 119206. https://doi.org/10.1016/j.bbamcr.2021.119206.
Xu, D., Shao, F., Bian, X., Meng, Y., Liang, T., & Lu, Z. (2021). The evolving landscape of noncanonical functions of metabolic enzymes in cancer and other pathologies. Cell Metabolism, 33(1), 33–50. https://doi.org/10.1016/j.cmet.2020.12.015.
Patra, K. C., Wang, Q., Bhaskar, P. T., Miller, L., Wang, Z., Wheaton, W., Chandel, N., Laakso, M., Muller, W. J., Allen, E. L., Jha, A. K., Smolen, G. A., Clasquin, M. F., Robey, B., & Hay, N. (2013). Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell, 24(2), 213–228. https://doi.org/10.1016/j.ccr.2013.06.014.
Rodríguez, A., De La Cera, T., Herrero, P., & Moreno, F. (2001). The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. The Biochemical Journal, 355(Pt 3), 625–631. https://doi.org/10.1042/bj3550625.
Vega, M., Riera, A., Fernández-Cid, A., Herrero, P., & Moreno, F. (2016). Hexokinase 2 is an intracellular glucose sensor of yeast cells that maintains the structure and activity of Mig1 protein repressor complex. The Journal of Biological Chemistry, 291(14), 7267–7285. https://doi.org/10.1074/jbc.M115.711408.
Papamichos-Chronakis, M., Gligoris, T., & Tzamarias, D. (2004). The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Reports, 5(4), 368–372. https://doi.org/10.1038/sj.embor.7400120.
Crozet, P., Margalha, L., Confraria, A., Rodrigues, A., Martinho, C., Adamo, M., Elias, C. A., & Baena-González, E. (2014). Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Frontiers in Plant Science, 5, 190. https://doi.org/10.3389/fpls.2014.00190.
Adeva-Andany, M., López-Ojén, M., Funcasta-Calderón, R., Ameneiros-Rodríguez, E., Donapetry-García, C., Vila-Altesor, M., & Rodríguez-Seijas, J. (2014). Comprehensive review on lactate metabolism in human health. Mitochondrion, 17, 76–100. https://doi.org/10.1016/j.mito.2014.05.007.
Fiume, L., Vettraino, M., Carnicelli, D., Arfilli, V., Di Stefano, G., & Brigotti, M. (2013). Galloflavin prevents the binding of lactate dehydrogenase A to single stranded DNA and inhibits RNA synthesis in cultured cells. Biochemical and Biophysical Research Communications, 430(2), 466–469. https://doi.org/10.1016/j.bbrc.2012.12.013.
Williams, K. R., Reddigari, S., & Patel, G. L. (1985). Identification of a nucleic acid helix-destabilizing protein from rat liver as lactate dehydrogenase-5. Proceedings of the National Academy of Sciences of the United States of America, 82(16), 5260–5264. https://doi.org/10.1073/pnas.82.16.5260.
Zheng, L., Roeder, R. G., & Luo, Y. (2003). S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell, 114(2), 255–266. https://doi.org/10.1016/s0092-8674(03)00552-x.
Fazioli, F., Minichiello, L., Matoska, V., Castagnino, P., Miki, T., Wong, W. T., & Di Fiore, P. P. (1993). Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. The EMBO Journal, 12(10), 3799–3808. https://doi.org/10.1002/j.1460-2075.1993.tb06058.x.
Cattaneo, A., Biocca, S., Corvaja, N., & Calissano, P. (1985). Nuclear localization of a lactic dehydrogenase with single-stranded DNA-binding properties. Experimental Cell Research, 161(1), 130–140. https://doi.org/10.1016/0014-4827(85)90497-5.
Zhong, X. H., & Howard, B. D. (1990). Phosphotyrosine-containing lactate dehydrogenase is restricted to the nuclei of PC12 pheochromocytoma cells. Molecular and Cellular Biology, 10(2), 770–776. https://doi.org/10.1128/mcb.10.2.770-776.1990.
Schultz, D. E., Hardin, C. C., & Lemon, S. M. (1996). Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5’-nontranslated RNA of hepatitis A virus. The Journal of Biological Chemistry, 271(24), 14134–14142. https://doi.org/10.1074/jbc.271.24.14134.
Pioli, P. A., Hamilton, B. J., Connolly, J. E., Brewer, G., & Rigby, W. F. (2002). Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. The Journal of Biological Chemistry, 277(38), 35738–35745. https://doi.org/10.1074/jbc.M204002200.
Andrade, J., Pearce, S. T., Zhao, H., & Barroso, M. (2004). Interactions among p22, glyceraldehyde-3-phosphate dehydrogenase and microtubules. The Biochemical Journal, 384(Pt 2), 327–336. https://doi.org/10.1042/BJ20040622.
Cancemi, P., Buttacavoli, M., Roz, E., & Feo, S. (2019). Expression of alpha-enolase (ENO1), Myc promoter-binding protein-1 (MBP-1) and matrix metalloproteinases (MMP-2 and MMP-9) reflect the nature and aggressiveness of breast tumors. International Journal of Molecular Sciences, 20(16), 3952. https://doi.org/10.3390/ijms20163952.
Chaudhary, D., & Miller, D. M. (1995). The c-myc promoter binding protein (MBP-1) and TBP bind simultaneously in the minor groove of the c-myc P2 promoter. Biochemistry, 34(10), 3438–3445. https://doi.org/10.1021/bi00010a036.
Ghosh, A. K., Steele, R., & Ray, R. B. (1999). MBP-1 physically associates with histone deacetylase for transcriptional repression. Biochemical and Biophysical Research Communications, 260(2), 405–409. https://doi.org/10.1006/bbrc.1999.0921.
Ghosh, A. K., Majumder, M., Steele, R., White, R. A., & Ray, R. B. (2001). A novel 16-kilodalton cellular protein physically interacts with and antagonizes the functional activity of c-myc promoter-binding protein 1. Molecular and Cellular Biology, 21(2), 655–662. https://doi.org/10.1128/MCB.21.2.655-662.2001.
Ogino, T., Yamadera, T., Nonaka, T., Imajoh-Ohmi, S., & Mizumoto, K. (2001). Enolase, a cellular glycolytic enzyme, is required for efficient transcription of Sendai virus genome. Biochemical and Biophysical Research Communications, 285(2), 447–455. https://doi.org/10.1006/bbrc.2001.5160.
Zahra, K., Dey, T., Ashish, Mishra, S. P., & Pandey, U. (2020). Pyruvate kinase M2 and cancer: The role of PKM2 in promoting tumorigenesis. Frontiers in Oncology, 10, 159. https://doi.org/10.3389/fonc.2020.00159.
Azoitei, N., Becher, A., Steinestel, K., Rouhi, A., Diepold, K., Genze, F., Simmet, T., & Seufferlein, T. (2016). PKM2 promotes tumor angiogenesis by regulating HIF-1α through NF-κB activation. Molecular Cancer, 15, 3. https://doi.org/10.1186/s12943-015-0490-2.
Demaria, M., & Poli, V. (2012). PKM2, STAT3 and HIF-1α: The Warburg’s vicious circle. JAK-STAT, 1(3), 194–196. https://doi.org/10.4161/jkst.20662.
Li, X., Zheng, Y., & Lu, Z. (2016). PGK1 is a new member of the protein kinome. Cell Cycle (Georgetown, Tex.), 15(14), 1803–1804. https://doi.org/10.1080/15384101.2016.1179037.
Yi, W., Clark, P. M., Mason, D. E., Keenan, M. C., Hill, C., Goddard, 3rd, W. A., Peters, E. C., Driggers, E. M., & Hsieh-Wilson, L. C. (2012). Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science (New York, N.Y.), 337(6097), 975–980. https://doi.org/10.1126/science.1222278.
Simula, L., Alifano, M., & Icard, P. (2022). How phosphofructokinase-1 promotes PI3K and YAP/TAZ in cancer: Therapeutic perspectives. Cancers, 14(10), 2478. https://doi.org/10.3390/cancers14102478.
Miller, J. L., & Grant, P. A. (2013). The role of DNA methylation and histone modifications in transcriptional regulation in humans. Sub-Cellular Biochemistry, 61, 289–317. https://doi.org/10.1007/978-94-007-4525-4_13.
Meier, J. L. (2013). Metabolic mechanisms of epigenetic regulation. ACS Chemical Biology, 8(12), 2607–2621. https://doi.org/10.1021/cb400689r.
Fan, J., Krautkramer, K. A., Feldman, J. L., & Denu, J. M. (2015). Metabolic regulation of histone post-translational modifications. ACS Chemical Biology, 10(1), 95–108. https://doi.org/10.1021/cb500846u.
Boon, R. (2021). Metabolic fuel for epigenetic: Nuclear production meets local consumption. Frontiers in Genetics, 12, 768996. https://doi.org/10.3389/fgene.2021.768996.
Sivanand, S., Viney, I., & Wellen, K. E. (2018). Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends in Biochemical Sciences, 43(1), 61–74. https://doi.org/10.1016/j.tibs.2017.11.004.
Kornacki, J. R., Stuparu, A. D., & Mrksich, M. (2015). Acetyltransferase p300/CBP associated Factor (PCAF) regulates crosstalk-dependent acetylation of histone H3 by distal site recognition. ACS Chemical Biology, 10(1), 157–164. https://doi.org/10.1021/cb5004527.
Evans, C. G., Chang, L., & Gestwicki, J. E. (2010). Heat shock protein 70 (hsp70) as an emerging drug target. Journal of Medicinal Chemistry, 53(12), 4585–4602. https://doi.org/10.1021/jm100054f.
Zhao, Z., & Shilatifard, A. (2019). Epigenetic modifications of histones in cancer. Genome Biology, 20(1), 245. https://doi.org/10.1186/s13059-019-1870-5.
Friis, R. M., Wu, B. P., Reinke, S. N., Hockman, D. J., Sykes, B. D., & Schultz, M. C. (2009). A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Research, 37(12), 3969–3980. https://doi.org/10.1093/nar/gkp270.
Hellemann, E., Walker, J. L., Lesko, M. A., Chandrashekarappa, D. G., Schmidt, M. C., O’Donnell, A. F., & Durrant, J. D. (2022). Novel mutation in hexokinase 2 confers resistance to 2-deoxyglucose by altering protein dynamics. PLoS Computational Biology, 18(3), e1009929. https://doi.org/10.1371/journal.pcbi.1009929.
Cheng, T., Sudderth, J., Yang, C., Mullen, A. R., Jin, E. S., Matés, J. M., & DeBerardinis, R. J. (2011). Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proceedings of the National Academy of Sciences of the United States of America, 108(21), 8674–8679. https://doi.org/10.1073/pnas.1016627108.
Ucar, D., Hu, Q., & Tan, K. (2011). Combinatorial chromatin modification patterns in the human genome revealed by subspace clustering. Nucleic Acids Research, 39(10), 4063–4075. https://doi.org/10.1093/nar/gkr016.
Matsuda, S., Adachi, J., Ihara, M., Tanuma, N., Shima, H., Kakizuka, A., Ikura, M., Ikura, T., & Matsuda, T. (2016). Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic Acids Research, 44(2), 636–647. https://doi.org/10.1093/nar/gkv967.
Latham, T., Mackay, L., Sproul, D., Karim, M., Culley, J., Harrison, D. J., Hayward, L., Langridge-Smith, P., Gilbert, N., & Ramsahoye, B. H. (2012). Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Research, 40(11), 4794–4803. https://doi.org/10.1093/nar/gks066.
Watanabe, H., Inaba, Y., Kimura, K., Matsumoto, M., Kaneko, S., Kasuga, M., & Inoue, H. (2018). Sirt2 facilitates hepatic glucose uptake by deacetylating glucokinase regulatory protein. Nature Communications, 9(1), 30. https://doi.org/10.1038/s41467-017-02537-6.
Cantó, C., Gerhart-Hines, Z., Feige, J. N., Lagouge, M., Noriega, L., Milne, J. C., Elliott, P. J., Puigserver, P., & Auwerx, J. (2009). AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature, 458(7241), 1056–1060. https://doi.org/10.1038/nature07813.
Yang, W., Xia, Y., Hawke, D., Li, X., Liang, J., Xing, D., Aldape, K., Hunter, T., Alfred Yung, W. K., & Lu, Z. (2012). PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell, 150(4), 685–696. https://doi.org/10.1016/j.cell.2012.07.018.
Yu, Q., Tong, C., Luo, M., Xue, X., Mei, Q., Ma, L., Yu, X., Mao, W., Kong, L., Yu, X., & Li, S. (2017). Regulation of SESAME-mediated H3T11 phosphorylation by glycolytic enzymes and metabolites. PloS One, 12(4), e0175576. https://doi.org/10.1371/journal.pone.0175576.
He, H., Lee, M. C., Zheng, L. L., Zheng, L., & Luo, Y. (2013). Integration of the metabolic/redox state, histone gene switching, DNA replication and S-phase progression by moonlighting metabolic enzymes. Bioscience Reports, 33(2), e00018. https://doi.org/10.1042/BSR20120059.
Cai, L., Sutter, B. M., Li, B., & Tu, B. P. (2011). Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular Cell, 42(4), 426–437. https://doi.org/10.1016/j.molcel.2011.05.004.
Lozoya, O. A., Wang, T., Grenet, D., Wolfgang, T. C., Sobhany, M., Ganini da Silva, D., Riadi, G., Chandel, N., Woychik, R. P., & Santos, J. H. (2019). Mitochondrial acetyl-CoA reversibly regulates locus-specific histone acetylation and gene expression. Life Science Alliance, 2(1), e201800228. https://doi.org/10.26508/lsa.201800228.
Ma, R., Wu, Y., Li, S., & Yu, X. (2021). Interplay between glucose metabolism and chromatin modifications in cancer. Frontiers in Cell and Developmental Biology, 9, 654337. https://doi.org/10.3389/fcell.2021.654337.
Keller, K. E., Doctor, Z. M., Dwyer, Z. W., & Lee, Y. S. (2014). SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Molecular Cell, 53(5), 700–709. https://doi.org/10.1016/j.molcel.2014.02.015.
Rosa, F., & Osorio, J. S. (2020). Quantitative determination of histone methylation via fluorescence resonance energy transfer (FRET) technology in immortalized bovine mammary alveolar epithelial cells supplemented with methionine. PloS One, 15(12), e0244135. https://doi.org/10.1371/journal.pone.0244135.
Amelio, I., Cutruzzolá, F., Antonov, A., Agostini, M., & Melino, G. (2014). Serine and glycine metabolism in cancer. Trends in Biochemical Sciences, 39(4), 191–198. https://doi.org/10.1016/j.tibs.2014.02.004.
Ghergurovich, J. M., Xu, X., Wang, J. Z., Yang, L., Ryseck, R. P., Wang, L., & Rabinowitz, J. D. (2021). Methionine synthase supports tumour tetrahydrofolate pools. Nature Metabolism, 3(11), 1512–1520. https://doi.org/10.1038/s42255-021-00465-w.
Liu, M., Saha, N., Gajan, A., Saadat, N., Gupta, S. V., & Pile, L. A. (2020). A complex interplay between SAM synthetase and the epigenetic regulator SIN3 controls metabolism and transcription. The Journal of Biological Chemistry, 295(2), 375–389. https://doi.org/10.1074/jbc.RA119.010032.
Wong, C. C., Qian, Y., & Yu, J. (2017). Interplay between epigenetics and metabolism in oncogenesis: Mechanisms and therapeutic approaches. Oncogene, 36(24), 3359–3374. https://doi.org/10.1038/onc.2016.485.
Gatla, H. R., Muniraj, N., Thevkar, P., Yavvari, S., Sukhavasi, S., & Makena, M. R. (2019). Regulation of chemokines and cytokines by histone deacetylases and an update on histone decetylase inhibitors in human diseases. International Journal of Molecular Sciences, 20(5), 1110. https://doi.org/10.3390/ijms20051110.
Das, P., & Taube, J. H. (2020). Regulating methylation at H3K27: A trick or treat for cancer cell plasticity. Cancers, 12(10), 2792. https://doi.org/10.3390/cancers12102792.
Soto-Heredero, G., Gómez de Las Heras, M. M., Gabandé-Rodríguez, E., Oller, J., & Mittelbrunn, M. (2020). Glycolysis - a key player in the inflammatory response. The FEBS Journal, 287(16), 3350–3369. https://doi.org/10.1111/febs.15327.
Dinarello, C. A. (2018). Overview of the IL-1 family in innate inflammation and acquired immunity. Immunological Reviews, 281(1), 8–27. https://doi.org/10.1111/imr.12621.
Sutinen, E. M., Pirttilä, T., Anderson, G., Salminen, A., & Ojala, J. O. (2012). Pro-inflammatory interleukin-18 increases Alzheimer’s disease-associated amyloid-β production in human neuron-like cells. Journal of Neuroinflammation, 9, 199. https://doi.org/10.1186/1742-2094-9-199.
Missiroli, S., Genovese, I., Perrone, M., Vezzani, B., Vitto, V. A. M., & Giorgi, C. (2020). The role of mitochondria in inflammation: From cancer to neurodegenerative disorders. Journal of Clinical Medicine, 9(3), 740. https://doi.org/10.3390/jcm9030740.
Bhattacharya, S., Ploplis, V. A., & Castellino, F. J. (2012). Bacterial plasminogen receptors utilize host plasminogen system for effective invasion and dissemination. Journal of Biomedicine & Biotechnology, 2012, 482096. https://doi.org/10.1155/2012/482096.
Yatsenko, T., Skrypnyk, M., Troyanovska, O., Tobita, M., Osada, T., Takahashi, S., Hattori, K., & Heissig, B. (2023). The role of the plasminogen/plasmin system in inflammation of the oral cavity. Cells, 12(3), 445. https://doi.org/10.3390/cells12030445.
Tristan, C., Shahani, N., Sedlak, T. W., & Sawa, A. (2011). The diverse functions of GAPDH: Views from different subcellular compartments. Cellular Signalling, 23(2), 317–323. https://doi.org/10.1016/j.cellsig.2010.08.003.
Liao, S. T., Han, C., Xu, D. Q., Fu, X. W., Wang, J. S., & Kong, L. Y. (2019). 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nature Communications, 10(1), 5091. https://doi.org/10.1038/s41467-019-13078-5.
Millet, P., Vachharajani, V., McPhail, L., Yoza, B., & McCall, C. E. (2016). GAPDH Binding to TNF-α mRNA Contributes to Posttranscriptional Repression in Monocytes: A Novel Mechanism of Communication between Inflammation and Metabolism. Journal of Immunology (Baltimore, Md.: 1950), 196(6), 2541–2551. https://doi.org/10.4049/jimmunol.1501345.
Lo Presti, M., Ferro, A., Contino, F., Mazzarella, C., Sbacchi, S., Roz, E., Lupo, C., Perconti, G., Giallongo, A., Migliorini, P., Marrazzo, A., & Feo, S. (2010). Myc promoter-binding protein-1 (MBP-1) is a novel potential prognostic marker in invasive ductal breast carcinoma. PloS One, 5(9), e12961. https://doi.org/10.1371/journal.pone.0012961.
Miller, D. M., Thomas, S. D., Islam, A., Muench, D., & Sedoris, K. (2012). c-Myc and cancer metabolism. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 18(20), 5546–5553. https://doi.org/10.1158/1078-0432.CCR-12-0977.
Sedoris, K. C., Thomas, S. D., & Miller, D. M. (2010). Hypoxia induces differential translation of enolase/MBP-1. BMC Cancer, 10, 157. https://doi.org/10.1186/1471-2407-10-157.
Goverman, J. (2009). Autoimmune T cell responses in the central nervous system. Nature Reviews Immunology, 9(6), 393–407. https://doi.org/10.1038/nri2550.
Liu, Z., Zhang, A., Zheng, L., Johnathan, A. F., Zhang, J., & Zhang, G. (2018). The biological significance and regulatory mechanism of c-Myc binding protein 1 (MBP-1). International Journal of Molecular Sciences, 19(12), 3868. https://doi.org/10.3390/ijms19123868.
Huang, C. K., Sun, Y., Lv, L., & Ping, Y. (2022). ENO1 and cancer. Molecular Therapy Oncolytics, 24, 288–298. https://doi.org/10.1016/j.omto.2021.12.026.
Su, Q., Tao, T., Tang, L., Deng, J., Darko, K. O., Zhou, S., Peng, M., He, S., Zeng, Q., Chen, A. F., & Yang, X. (2018). Down-regulation of PKM2 enhances anticancer efficiency of THP on bladder cancer. Journal of Cellular and Molecular Medicine, 22(5), 2774–2790. https://doi.org/10.1111/jcmm.13571.
Gupta, V., & Bamezai, R. N. (2010). Human pyruvate kinase M2: A multifunctional protein. Protein Science: A Publication of the Protein Society, 19(11), 2031–2044. https://doi.org/10.1002/pro.505.
Corcoran, S. E., & O’Neill, L. A. (2016). HIF1α and metabolic reprogramming in inflammation. The Journal of Clinical Investigation, 126(10), 3699–3707. https://doi.org/10.1172/JCI84431.
Dietrich, C., & Kaina, B. (2010). The aryl hydrocarbon receptor (AhR) in the regulation of cell-cell contact and tumor growth. Carcinogenesis, 31(8), 1319–1328. https://doi.org/10.1093/carcin/bgq028.
Israelsen, W. J., & Vander Heiden, M. G. (2015). Pyruvate kinase: Function, regulation and role in cancer. Seminars in Cell & Developmental Biology, 43, 43–51. https://doi.org/10.1016/j.semcdb.2015.08.004.
Tang, Y., Gu, S., Zhu, L., Wu, Y., Zhang, W., & Zhao, C. (2022). LDHA: The Obstacle to T cell responses against tumor. Frontiers in Oncology, 12, 1036477. https://doi.org/10.3389/fonc.2022.1036477.
Van Wilpe, S., Koornstra, R., Den Brok, M., De Groot, J. W., Blank, C., De Vries, J., Gerritsen, W., & Mehra, N. (2020). Lactate dehydrogenase: A marker of diminished antitumor immunity. Oncoimmunology, 9(1), 1731942. https://doi.org/10.1080/2162402X.2020.1731942.
Serra, M., Di Matteo, M., Serneels, J., Pal, R., Cafarello, S. T., Lanza, M., Sanchez-Martin, C., Evert, M., Castegna, A., Calvisi, D. F., Mazzone, M., & Columbano, A. (2022). Deletion of lactate dehydrogenase-A impairs oncogene-induced mouse hepatocellular carcinoma development. Cellular and Molecular Gastroenterology and Hepatology, 14(3), 609–624. https://doi.org/10.1016/j.jcmgh.2022.06.003.
Chen, J., Adamiak, W., Huang, G., Atasoy, U., Rostami, A., & Yu, S. (2017). Interaction of RNA-binding protein HuR and miR-466i regulates GM-CSF expression. Scientific Reports, 7(1), 17233. https://doi.org/10.1038/s41598-017-17371-5.
Elmore, S. (2007). Apoptosis: A review of programmed cell death. Toxicologic Pathology, 35(4), 495–516. https://doi.org/10.1080/01926230701320337.
Mason, E. F., & Rathmell, J. C. (2011). Cell metabolism: An essential link between cell growth and apoptosis. Biochimica et Biophysica Acta, 1813(4), 645–654. https://doi.org/10.1016/j.bbamcr.2010.08.011.
Schoeniger, A., Wolf, P., & Edlich, F. (2022). How do hexokinases inhibit receptor-mediated apoptosis? Biology, 11(3), 412. https://doi.org/10.3390/biology11030412.
Garrido, C., Galluzzi, L., Brunet, M., Puig, P. E., Didelot, C., & Kroemer, G. (2006). Mechanisms of cytochrome c release from mitochondria. Cell Death and Differentiation, 13(9), 1423–1433. https://doi.org/10.1038/sj.cdd.4401950.
Szlyk, B., Braun, C. R., Ljubicic, S., Patton, E., Bird, G. H., Osundiji, M. A., Matschinsky, F. M., Walensky, L. D., & Danial, N. N. (2014). A phospho-BAD BH3 helix activates glucokinase by a mechanism distinct from that of allosteric activators. Nature Structural & Molecular Biology, 21(1), 36–42. https://doi.org/10.1038/nsmb.2717.
Jahan, I., Corbin, K. L., Bogart, A. M., Whitticar, N. B., Waters, C. D., Schildmeyer, C., Vann, N. W., West, H. L., Law, N. C., Wiseman, J. S., & Nunemaker, C. S. (2018). Reducing glucokinase activity restores endogenous pulsatility and enhances insulin secretion in Islets From db/db mice. Endocrinology, 159(11), 3747–3760. https://doi.org/10.1210/en.2018-00589.
White, M. R., & Garcin, E. D. (2016). The sweet side of RNA regulation: Glyceraldehyde-3-phosphate dehydrogenase as a noncanonical RNA-binding protein. Wiley interdisciplinary reviews. RNA, 7(1), 53–70. https://doi.org/10.1002/wrna.1315.
Chuang, D. M., Hough, C., & Senatorov, V. V. (2005). Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annual Review of Pharmacology and Toxicology, 45, 269–290. https://doi.org/10.1146/annurev.pharmtox.45.120403.095902.
Shashidharan, P., Chalmers-Redman, R. M., Carlile, G. W., Rodic, V., Gurvich, N., Yuen, T., Tatton, W. G., & Sealfon, S. C. (1999). Nuclear translocation of GAPDH-GFP fusion protein during apoptosis. Neuroreport, 10(5), 1149–1153. https://doi.org/10.1097/00001756-199904060-00045.
Gibson, R. M. (2001). Does apoptosis have a role in neurodegeneration? BMJ (Clinical research ed.), 322(7301), 1539–1540. https://doi.org/10.1136/bmj.322.7301.1539.
El Kadmiri, N., Slassi, I., El Moutawakil, B., Nadifi, S., Tadevosyan, A., Hachem, A., & Soukri, A. (2014). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer’s disease. Pathologie-Biologie, 62(6), 333–336. https://doi.org/10.1016/j.patbio.2014.08.002.
Mikhaylova, E. R., Lazarev, V. F., Nikotina, A. D., Margulis, B. A., & Guzhova, I. V. (2016). Glyceraldehyde 3-phosphate dehydrogenase augments the intercellular transmission and toxicity of polyglutamine aggregates in a cell model of Huntington disease. Journal of Neurochemistry, 136(5), 1052–1063. https://doi.org/10.1111/jnc.13463.
Schulz, L. C., & Bahr, J. M. (2003). Glucose-6-phosphate isomerase is necessary for embryo implantation in the domestic ferret. Proceedings of the National Academy of Sciences of the United States of America, 100(14), 8561–8566. https://doi.org/10.1073/pnas.1531024100.
Coulam, C. B., Kaider, B. D., Kaider, A. S., Janowicz, P., & Roussev, R. G. (1997). Antiphospholipid antibodies associated with implantation failure after IVF/ET. Journal of Assisted Reproduction and Genetics, 14(10), 603–608. https://doi.org/10.1023/a:1022588903620.
Li, Y., Jia, Y., Che, Q., Zhou, Q., Wang, K., & Wan, X. P. (2015). AMF/PGI-mediated tumorigenesis through MAPK-ERK signaling in endometrial carcinoma. Oncotarget, 6(28), 26373–26387. https://doi.org/10.18632/oncotarget.4708.
Nakajima, K., & Raz, A. (2020). Autocrine motility factor and its receptor expression in musculoskeletal tumors. Journal of Bone Oncology, 24, 100318. https://doi.org/10.1016/j.jbo.2020.100318.
Lucarelli, G., Rutigliano, M., Sanguedolce, F., Galleggiante, V., Giglio, A., Cagiano, S., Bufo, P., Maiorano, E., Ribatti, D., Ranieri, E., Gigante, M., Gesualdo, L., Ferro, M., de Cobelli, O., Buonerba, C., Di Lorenzo, G., De Placido, S., Palazzo, S., Bettocchi, C., Ditonno, P., & Battaglia, M. (2015). Increased expression of the autocrine motility factor is associated with poor prognosis in patients with clear cell-renal cell carcinoma. Medicine, 94(46), e2117. https://doi.org/10.1097/MD.0000000000002117.
Khajah, M. A., Khushaish, S., & Luqmani, Y. A. (2021). Lactate dehydrogenase A or B knockdown reduces lactate production and inhibits breast cancer cell motility in vitro. Frontiers in Pharmacology, 12, 747001. https://doi.org/10.3389/fphar.2021.747001.
Parzych, K. R., & Klionsky, D. J. (2014). An overview of autophagy: Morphology, mechanism, and regulation. Antioxidants & Redox Signaling, 20(3), 460–473. https://doi.org/10.1089/ars.2013.5371.
Kroemer, G., Mariño, G., & Levine, B. (2010). Autophagy and the integrated stress response. Molecular Cell, 40(2), 280–293. https://doi.org/10.1016/j.molcel.2010.09.023.
Khandia, R., Dadar, M., Munjal, A., Dhama, K., Karthik, K., Tiwari, R., Yatoo, M. I., Iqbal, H. M. N., Singh, K. P., Joshi, S. K., & Chaicumpa, W. (2019). A comprehensive review of autophagy and its various roles in infectious, non-infectious, and lifestyle diseases: Current knowledge and prospects for disease prevention, novel drug design, and therapy. Cells, 8(7), 674. https://doi.org/10.3390/cells8070674.
Yun, C. W., & Lee, S. H. (2018). The roles of autophagy in cancer. International Journal of Molecular Sciences, 19(11), 3466. https://doi.org/10.3390/ijms19113466.
Srinivas, U. S., Tan, B. W. Q., Vellayappan, B. A., & Jeyasekharan, A. D. (2019). ROS and the DNA damage response in cancer. Redox Biology, 25, 101084. https://doi.org/10.1016/j.redox.2018.101084.
Zheng, J. (2012). Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncology Letters, 4(6), 1151–1157. https://doi.org/10.3892/ol.2012.928.
Wang, X., Lu, S., He, C., Wang, C., Wang, L., Piao, M., Chi, G., Luo, Y., & Ge, P. (2019). RSL3 induced autophagic death in glioma cells via causing glycolysis dysfunction. Biochemical and Biophysical Research Communications, 518(3), 590–597. https://doi.org/10.1016/j.bbrc.2019.08.096.
Kim, J. H., Nam, B., Choi, Y. J., Kim, S. Y., Lee, J. E., Sung, K. J., Kim, W. S., Choi, C. M., Chang, E. J., Koh, J. S., Song, J. S., Yoon, S., Lee, J. C., Rho, J. K., & Son, J. (2018). Enhanced glycolysis supports cell survival in EGFR-mutant lung adenocarcinoma by inhibiting autophagy-mediated EGFR degradation. Cancer Research, 78(16), 4482–4496. https://doi.org/10.1158/0008-5472.CAN-18-0117.
Chu, Y., Chang, Y., Lu, W., Sheng, X., Wang, S., Xu, H., & Ma, J. (2020). Regulation of autophagy by glycolysis in cancer. Cancer Management and Research, 12, 13259–13271. https://doi.org/10.2147/CMAR.S279672.
Kothari, S. S., Abrahamsen, M. S., Cole, T., & Hammond, W. P. (1995). Expression of granulocyte colony stimulating factor (G-CSF) and granulocyte/macrophage colony stimulating factor (GM-CSF) mRNA upon stimulation with phorbol ester. Blood Cells, Molecules & Diseases, 21(3), 192–200. https://doi.org/10.1006/bcmd.1995.0022.
Zhang, X. Y., Zhang, M., Cong, Q., Zhang, M. X., Zhang, M. Y., Lu, Y. Y., & Xu, C. J. (2018). Hexokinase 2 confers resistance to cisplatin in ovarian cancer cells by enhancing cisplatin-induced autophagy. The International Journal of Biochemistry & Cell Biology, 95, 9–16. https://doi.org/10.1016/j.biocel.2017.12.010.
Tan, W. X., Xu, T. M., Zhou, Z. L., Lv, X. J., Liu, J., Zhang, W. J., & Cui, M. H. (2019). TRP14 promotes resistance to cisplatin by inducing autophagy in ovarian cancer. Oncology Reports, 42(4), 1343–1354. https://doi.org/10.3892/or.2019.7258.
Pajak, B., Siwiak, E., Sołtyka, M., Priebe, A., Zieliński, R., Fokt, I., Ziemniak, M., Jaśkiewicz, A., Borowski, R., Domoradzki, T., & Priebe, W. (2019). 2-Deoxy-d-Glucose and its analogs: From diagnostic to therapeutic agents. International Journal of Molecular Sciences, 21(1), 234. https://doi.org/10.3390/ijms21010234.
Brohée, L., Peulen, O., Nusgens, B., Castronovo, V., Thiry, M., Colige, A. C., & Deroanne, C. F. (2018). Propranolol sensitizes prostate cancer cells to glucose metabolism inhibition and prevents cancer progression. Scientific Reports, 8(1), 7050. https://doi.org/10.1038/s41598-018-25340-9.
Ye, M., Wang, S., Wan, T., Jiang, R., Qiu, Y., Pei, L., Pang, N., Huang, Y., Huang, Y., Zhang, Z., & Yang, L. (2017). Combined inhibitions of glycolysis and AKT/autophagy can overcome resistance to EGFR-targeted therapy of lung cancer. Journal of Cancer, 8(18), 3774–3784. https://doi.org/10.7150/jca.21035.
Herzig, S., & Shaw, R. J. (2018). AMPK: Guardian of metabolism and mitochondrial homeostasis. Nature Reviews Molecular Cell Biology, 19(2), 121–135. https://doi.org/10.1038/nrm.2017.95.
Chang, C., Su, H., Zhang, D., Wang, Y., Shen, Q., Liu, B., Huang, R., Zhou, T., Peng, C., Wong, C. C., Shen, H. M., Lippincott-Schwartz, J., & Liu, W. (2015). AMPK-dependent phosphorylation of GAPDH Triggers Sirt1 activation and is necessary for autophagy upon glucose starvation. Molecular Cell, 60(6), 930–940. https://doi.org/10.1016/j.molcel.2015.10.037.
Butera, G., Mullappilly, N., Masetto, F., Palmieri, M., Scupoli, M. T., Pacchiana, R., & Donadelli, M. (2019). Regulation of autophagy by nuclear GAPDH and its aggregates in cancer and neurodegenerative disorders. International Journal of Molecular Sciences, 20(9), 2062. https://doi.org/10.3390/ijms20092062.
Chambers, A., Tsang, J. S., Stanway, C., Kingsman, A. J., & Kingsman, S. M. (1989). Transcriptional control of the Saccharomyces cerevisiae PGK gene by RAP1. Molecular and Cellular Biology, 9(12), 5516–5524. https://doi.org/10.1128/mcb.9.12.5516-5524.1989.
Park, J. M., Seo, M., Jung, C. H., Grunwald, D., Stone, M., Otto, N. M., Toso, E., Ahn, Y., Kyba, M., Griffin, T. J., Higgins, L., & Kim, D. H. (2018). ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy, 14(4), 584–597. https://doi.org/10.1080/15548627.2017.1422851.
Das, C. K., Parekh, A., Parida, P. K., Bhutia, S. K., & Mandal, M. (2019). Lactate dehydrogenase A regulates autophagy and tamoxifen resistance in breast cancer. Biochimica et Biophysica Acta. Molecular Cell Research, 1866(6), 1004–1018. https://doi.org/10.1016/j.bbamcr.2019.03.004.
Urbańska, K., & Orzechowski, A. (2019). Unappreciated role of LDHA and LDHB to control apoptosis and autophagy in tumor cells. International Journal Of Molecular Sciences, 20(9), 2085. https://doi.org/10.3390/ijms20092085.
Shi, L., Yan, H., An, S., Shen, M., Jia, W., Zhang, R., Zhao, L., Huang, G., & Liu, J. (2019). SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Molecular Oncology, 13(2), 358–375. https://doi.org/10.1002/1878-0261.12408.
Kim, L. C., Cook, R. S., & Chen, J. (2017). mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene, 36(16), 2191–2201. https://doi.org/10.1038/onc.2016.363.
Lv, D., Guo, L., Zhang, T., & Huang, L. (2017). PRAS40 signaling in tumor. Oncotarget, 8(40), 69076–69085. https://doi.org/10.18632/oncotarget.17299.
He, X., Du, S., Lei, T., Li, X., Liu, Y., Wang, H., Tong, R., & Wang, Y. (2017). PKM2 in carcinogenesis and oncotherapy. Oncotarget, 8(66), 110656–110670. https://doi.org/10.18632/oncotarget.22529.
Magaway, C., Kim, E., & Jacinto, E. (2019). Targeting mTOR and metabolism in cancer: Lessons and innovations. Cells, 8(12), 1584. https://doi.org/10.3390/cells8121584.
Batlle, E., & Massagué, J. (2019). Transforming growth Factor-β signaling in immunity and cancer. Immunity, 50(4), 924–940. https://doi.org/10.1016/j.immuni.2019.03.024.
Parker, T. W., & Neufeld, K. L. (2020). APC controls Wnt-induced β-catenin destruction complex recruitment in human colonocytes. Scientific Reports, 10(1), 2957. https://doi.org/10.1038/s41598-020-59899-z.
Vallée, A., Lecarpentier, Y., & Vallée, J. N. (2021). The key role of the WNT/β-Catenin pathway in metabolic reprogramming in cancers under normoxic conditions. Cancers, 13(21), 5557. https://doi.org/10.3390/cancers13215557.
Pate, K. T., Stringari, C., Sprowl-Tanio, S., Wang, K., TeSlaa, T., Hoverter, N. P., McQuade, M. M., Garner, C., Digman, M. A., Teitell, M. A., Edwards, R. A., Gratton, E., & Waterman, M. L. (2014). Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. The EMBO Journal, 33(13), 1454–1473. https://doi.org/10.15252/embj.201488598.
Pérez-Tomás, R., & Pérez-Guillén, I. (2020). Lactate in the tumor microenvironment: An essential molecule in cancer progression and treatment. Cancers, 12(11), 3244. https://doi.org/10.3390/cancers12113244.
Mo, Y., Wang, Y., Zhang, L., Yang, L., Zhou, M., Li, X., Li, Y., Li, G., Zeng, Z., Xiong, W., Xiong, F., & Guo, C. (2019). The role of Wnt signaling pathway in tumor metabolic reprogramming. Journal of Cancer, 10(16), 3789–3797. https://doi.org/10.7150/jca.31166.
Yoo, H. C., Yu, Y. C., Sung, Y., & Han, J. M. (2020). Glutamine reliance in cell metabolism. Experimental & Molecular Medicine, 52(9), 1496–1516. https://doi.org/10.1038/s12276-020-00504-8.
Li, S., Liu, F., Xu, L., Li, C., Yang, X., Guo, B., Gu, J., & Wang, L. (2020). Wnt/β-catenin signaling axis is required for TFEB-mediated gastric cancer metastasis and epithelial-mesenchymal transition. Molecular Cancer Research: MCR, 18(11), 1650–1659. https://doi.org/10.1158/1541-7786.MCR-20-0180.
Shang, S., Hua, F., & Hu, Z. W. (2017). The regulation of β-catenin activity and function in cancer: Therapeutic opportunities. Oncotarget, 8(20), 33972–33989. https://doi.org/10.18632/oncotarget.15687.
Long, Y. C., & Zierath, J. R. (2006). AMP-activated protein kinase signaling in metabolic regulation. The Journal of Clinical Investigation, 116(7), 1776–1783. https://doi.org/10.1172/JCI29044.
Panieri, E., & Santoro, M. M. (2016). ROS homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death & Disease, 7(6), e2253. https://doi.org/10.1038/cddis.2016.105.
Gorrini, C., Harris, I. S., & Mak, T. W. (2013). Modulation of oxidative stress as an anticancer strategy. Nature Reviews Drug Discovery, 12(12), 931–947. https://doi.org/10.1038/nrd4002.
Weinberg, F., Hamanaka, R., Wheaton, W. W., Weinberg, S., Joseph, J., Lopez, M., Kalyanaraman, B., Mutlu, G. M., Budinger, G. R., & Chandel, N. S. (2010). Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America, 107(19), 8788–8793. https://doi.org/10.1073/pnas.1003428107.
Zu, X. L., & Guppy, M. (2004). Cancer metabolism: Facts, fantasy, and fiction. Biochemical and Biophysical Research Communications, 313(3), 459–465. https://doi.org/10.1016/j.bbrc.2003.11.136.
Liberti, M. V., & Locasale, J. W. (2016). The Warburg effect: How does it benefit cancer cells? Trends in Biochemical Sciences, 41(3), 211–218. https://doi.org/10.1016/j.tibs.2015.12.001.
Patra, K. C., & Hay, N. (2014). The pentose phosphate pathway and cancer. Trends in Biochemical Sciences, 39(8), 347–354. https://doi.org/10.1016/j.tibs.2014.06.005.
Hardie, D. G. (2011). AMP-activated protein kinase: An energy sensor that regulates all aspects of cell function. Genes & Development, 25(18), 1895–1908. https://doi.org/10.1101/gad.17420111.
Garcia, D., & Shaw, R. J. (2017). AMPK: Mechanisms of cellular energy sensing and restoration of metabolic balance. Molecular Cell, 66(6), 789–800. https://doi.org/10.1016/j.molcel.2017.05.032.
Shackelford, D. B., & Shaw, R. J. (2009). The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nature Reviews Cancer, 9(8), 563–575. https://doi.org/10.1038/nrc2676.
Sun, S. Y. (2010). N-acetylcysteine, reactive oxygen species and beyond. Cancer Biology & Therapy, 9(2), 109–110. https://doi.org/10.4161/cbt.9.2.10583.
Jeon, S. M., Chandel, N. S., & Hay, N. (2012). AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature, 485(7400), 661–665. https://doi.org/10.1038/nature11066.
Houten, S. M., & Wanders, R. J. (2010). A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. Journal of Inherited Metabolic Disease, 33(5), 469–477. https://doi.org/10.1007/s10545-010-9061-2.
Rabinovitch, R. C., Samborska, B., Faubert, B., Ma, E. H., Gravel, S. P., Andrzejewski, S., Raissi, T. C., Pause, A., St-Pierre, J., & Jones, R. G. (2017). AMPK Maintains cellular metabolic homeostasis through regulation of mitochondrial reactive oxygen species. Cell Reports, 21(1), 1–9. https://doi.org/10.1016/j.celrep.2017.09.026.
Marin, T. L., Gongol, B., Zhang, F., Martin, M., Johnson, D. A., Xiao, H., Wang, Y., Subramaniam, S., Chien, S., & Shyy, J. Y. (2017). AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Science Signaling, 10(464), eaaf7478. https://doi.org/10.1126/scisignal.aaf7478.
Zimmermann, K., Baldinger, J., Mayerhofer, B., Atanasov, A. G., Dirsch, V. M., & Heiss, E. H. (2015). Activated AMPK boosts the Nrf2/HO-1 signaling axis-A role for the unfolded protein response. Free Radical Biology & Medicine, 88(Pt B), 417–426. https://doi.org/10.1016/j.freeradbiomed.2015.03.030.
Kobayashi, A., Kang, M. I., Okawa, H., Ohtsuji, M., Zenke, Y., Chiba, T., Igarashi, K., & Yamamoto, M. (2004). Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Molecular and Cellular Biology, 24(16), 7130–7139. https://doi.org/10.1128/MCB.24.16.7130-7139.2004.
Ma, Q. (2013). Role of nrf2 in oxidative stress and toxicity. Annual Review of Pharmacology and Toxicology, 53, 401–426. https://doi.org/10.1146/annurev-pharmtox-011112-140320.
Zambrano, A., Molt, M., Uribe, E., & Salas, M. (2019). Glut 1 in cancer cells and the inhibitory action of resveratrol as a potential therapeutic strategy. International Journal of Molecular Sciences, 20(13), 3374. https://doi.org/10.3390/ijms20133374.
Ancey, P. B., Contat, C., & Meylan, E. (2018). Glucose transporters in cancer - from tumor cells to the tumor microenvironment. The FEBS Journal, 285(16), 2926–2943. https://doi.org/10.1111/febs.14577.
Lee, E. E., Ma, J., Sacharidou, A., Mi, W., Salato, V. K., Nguyen, N., Jiang, Y., Pascual, J. M., North, P. E., Shaul, P. W., Mettlen, M., & Wang, R. C. (2015). A protein kinase C phosphorylation motif in GLUT1 affects glucose transport and is mutated in GLUT1 deficiency syndrome. Molecular Cell, 58(5), 845–853. https://doi.org/10.1016/j.molcel.2015.04.015.
Romero-Garcia, S., Lopez-Gonzalez, J. S., Báez-Viveros, J. L., Aguilar-Cazares, D., & Prado-Garcia, H. (2011). Tumor cell metabolism: An integral view. Cancer Biology & Therapy, 12(11), 939–948. https://doi.org/10.4161/cbt.12.11.18140.
Stincone, A., Prigione, A., Cramer, T., Wamelink, M. M., Campbell, K., Cheung, E., Olin-Sandoval, V., Grüning, N. M., Krüger, A., Tauqeer Alam, M., Keller, M. A., Breitenbach, M., Brindle, K. M., Rabinowitz, J. D., & Ralser, M. (2015). The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biological Reviews of the Cambridge Philosophical Society, 90(3), 927–963. https://doi.org/10.1111/brv.12140.
Shibuya, M. (2011). Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: A crucial target for anti- and pro-angiogenic therapies. Genes & Cancer, 2(12), 1097–1105. https://doi.org/10.1177/1947601911423031.
Estrella, V., Chen, T., Lloyd, M., Wojtkowiak, J., Cornnell, H. H., Ibrahim-Hashim, A., Bailey, K., Balagurunathan, Y., Rothberg, J. M., Sloane, B. F., Johnson, J., Gatenby, R. A., & Gillies, R. J. (2013). Acidity generated by the tumor microenvironment drives local invasion. Cancer Research, 73(5), 1524–1535. https://doi.org/10.1158/0008-5472.CAN-12-2796.
Brooks, G. A., Dubouchaud, H., Brown, M., Sicurello, J. P., & Butz, C. E. (1999). Role of mitochondrial lactate dehydrogenase and lactate oxidation in the intracellular lactate shuttle. Proceedings of the National Academy of Sciences of the United States of America, 96(3), 1129–1134. https://doi.org/10.1073/pnas.96.3.1129.
Lagarde, D., Jeanson, Y., Portais, J. C., Galinier, A., Ader, I., Casteilla, L., & Carrière, A. (2021). Lactate fluxes and plasticity of adipose tissues: A redox perspective. Frontiers in Physiology, 12, 689747. https://doi.org/10.3389/fphys.2021.689747.
Xie, N., Zhang, L., Gao, W., Huang, C., Huber, P. E., Zhou, X., Li, C., Shen, G., & Zou, B. (2020). NAD+ metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduction and Targeted Therapy, 5(1), 227. https://doi.org/10.1038/s41392-020-00311-7.
Go, S., Kramer, T. T., Verhoeven, A. J., Oude Elferink, R. P. J., & Chang, J. C. (2021). The extracellular lactate-to-pyruvate ratio modulates the sensitivity to oxidative stress-induced apoptosis via the cytosolic NADH/NAD+ redox state. Apoptosis: An International Journal on Programmed Cell Death, 26(1-2), 38–51. https://doi.org/10.1007/s10495-020-01648-8.
Svedružić, Ž. M., Odorčić, I., Chang, C. H., & Svedružić, D. (2020). Substrate channeling via a transient protein-protein complex: The case of D-Glyceraldehyde-3-phosphate dehydrogenase and L-lactate dehydrogenase. Scientific Reports, 10(1), 10404. https://doi.org/10.1038/s41598-020-67079-2.
Vijay, N., & Morris, M. E. (2014). Role of monocarboxylate transporters in drug delivery to the brain. Current Pharmaceutical Design, 20(10), 1487–1498. https://doi.org/10.2174/13816128113199990462.
Rabinowitz, J. D., & Enerbäck, S. (2020). Lactate: The ugly duckling of energy metabolism. Nature Metabolism, 2(7), 566–571. https://doi.org/10.1038/s42255-020-0243-4.
Brooks, G. A. (2018). The science and translation of lactate shuttle theory. Cell Metabolism, 27(4), 757–785. https://doi.org/10.1016/j.cmet.2018.03.008.
Martínez-Reyes, I., & Chandel, N. S. (2020). Mitochondrial TCA cycle metabolites control physiology and disease. Nature Communications, 11(1), 102. https://doi.org/10.1038/s41467-019-13668-3.
Hashimoto, T., Hussien, R., Cho, H. S., Kaufer, D., & Brooks, G. A. (2008). Evidence for the mitochondrial lactate oxidation complex in rat neurons: Demonstration of an essential component of brain lactate shuttles. PloS One, 3(8), e2915. https://doi.org/10.1371/journal.pone.0002915.
Lin, Y., Wang, Y., & Li, P. F. (2022). Mutual regulation of lactate dehydrogenase and redox robustness. Frontiers in Physiology, 13, 1038421. https://doi.org/10.3389/fphys.2022.1038421.
Li, X., Yang, Y., Zhang, B., Lin, X., Fu, X., An, Y., Zou, Y., Wang, J. X., Wang, Z., & Yu, T. (2022). Lactate metabolism in human health and disease. Signal Transduction and Targeted Therapy, 7(1), 305. https://doi.org/10.1038/s41392-022-01151-3.
Kane, D. A. (2014). Lactate oxidation at the mitochondria: A lactate-malate-aspartate shuttle at work. Frontiers in Neuroscience, 8, 366. https://doi.org/10.3389/fnins.2014.00366.
Peters, A. L., & Van Noorden, C. J. (2009). Glucose-6-phosphate dehydrogenase deficiency and malaria: Cytochemical detection of heterozygous G6PD deficiency in women. The Journal of Histochemistry and Cytochemistry: Official Journal of the Histochemistry Society, 57(11), 1003–1011. https://doi.org/10.1369/jhc.2009.953828.
Dey, S., Sidor, A., & O’Rourke, B. (2016). Compartment-specific control of reactive oxygen species scavenging by antioxidant pathway enzymes. The Journal of Biological Chemistry, 291(21), 11185–11197. https://doi.org/10.1074/jbc.M116.726968.
Polat, I. H., Tarrado-Castellarnau, M., Bharat, R., Perarnau, J., Benito, A., Cortés, R., Sabatier, P., & Cascante, M. (2021). Oxidative pentose phosphate pathway enzyme 6-phosphogluconate dehydrogenase plays a key role in breast cancer metabolism. Biology, 10(2), 85. https://doi.org/10.3390/biology10020085.
Szablewski, L. (2022). Glucose transporters as markers of diagnosis and prognosis in cancer diseases. Oncology Reviews, 16(1), 561. https://doi.org/10.4081/oncol.2022.561.
Stanirowski, P. J., Szukiewicz, D., Majewska, A., Wątroba, M., Pyzlak, M., Bomba-Opoń, D., & Wielgoś, M. (2022). Placental expression of glucose transporters GLUT-1, GLUT-3, GLUT-8 and GLUT-12 in pregnancies complicated by gestational and type 1 diabetes mellitus. Journal of Diabetes Investigation, 13(3), 560–570. https://doi.org/10.1111/jdi.13680.
Wu, N., Zheng, B., Shaywitz, A., Dagon, Y., Tower, C., Bellinger, G., Shen, C. H., Wen, J., Asara, J., McGraw, T. E., Kahn, B. B., & Cantley, L. C. (2013). AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Molecular Cell, 49(6), 1167–1175. https://doi.org/10.1016/j.molcel.2013.01.035.
Dahl, E. S., & Aird, K. M. (2017). Ataxia-Telangiectasia mutated modulation of carbon metabolism in cancer. Frontiers in Oncology, 7, 291. https://doi.org/10.3389/fonc.2017.00291.
Maréchal, A., & Zou, L. (2013). DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor Perspectives in Biology, 5(9), a012716. https://doi.org/10.1101/cshperspect.a012716.
Xie, X., Zhang, Y., Wang, Z., Wang, S., Jiang, X., Cui, H., Zhou, T., He, Z., Feng, H., Guo, Q., Song, X., & Cao, L. (2021). ATM at the crossroads of reactive oxygen species and autophagy. International Journal of Biological Sciences, 17(12), 3080–3090. https://doi.org/10.7150/ijbs.63963.
Kurz, E. U., Douglas, P., & Lees-Miller, S. P. (2004). Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. The Journal of Biological Chemistry, 279(51), 53272–53281. https://doi.org/10.1074/jbc.M406879200.
Park, J., Kim, J., & Mikami, T. (2021). Exercise-induced lactate release mediates mitochondrial biogenesis in the hippocampus of mice via monocarboxylate transporters. Frontiers in Physiology, 12, 736905. https://doi.org/10.3389/fphys.2021.736905.
Bhatti, M. S., & Frostig, R. D. (2023). Astrocyte-neuron lactate shuttle plays a pivotal role in sensory-based neuroprotection in a rat model of permanent middle cerebral artery occlusion. Scientific Reports, 13(1), 12799. https://doi.org/10.1038/s41598-023-39574-9.
Zorov, D. B., Juhaszova, M., & Sollott, S. J. (2014). Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiological Reviews, 94(3), 909–950. https://doi.org/10.1152/physrev.00026.2013.
Johri, A., & Beal, M. F. (2012). Mitochondrial dysfunction in neurodegenerative diseases. The Journal of Pharmacology and Experimental Therapeutics, 342(3), 619–630. https://doi.org/10.1124/jpet.112.192138.
Höhn, A., Tramutola, A., & Cascella, R. (2020). Proteostasis failure in neurodegenerative diseases: Focus on oxidative stress. Oxidative Medicine and Cellular Longevity, 2020, 5497046. https://doi.org/10.1155/2020/5497046.
Tauffenberger, A., Fiumelli, H., Almustafa, S., & Magistretti, P. J. (2019). Lactate and pyruvate promote oxidative stress resistance through hormetic ROS signaling. Cell Death & Disease, 10(9), 653. https://doi.org/10.1038/s41419-019-1877-6.
Zou, G. P., Wang, T., Xiao, J. X., Wang, X. Y., Jiang, L. P., Tou, F. F., Chen, Z. P., Qu, X. H., & Han, X. J. (2023). Lactate protects against oxidative stress-induced retinal degeneration by activating autophagy. Free Radical Biology & Medicine, 194, 209–219. https://doi.org/10.1016/j.freeradbiomed.2022.12.004.
Schiliro, C., & Firestein, B. L. (2021). Mechanisms of metabolic reprogramming in cancer cells supporting enhanced growth and proliferation. Cells, 10(5), 1056. https://doi.org/10.3390/cells10051056.
Cho, E. S., Cha, Y. H., Kim, H. S., Kim, N. H., & Yook, J. I. (2018). The pentose phosphate pathway as a potential target for cancer therapy. Biomolecules & Therapeutics, 26(1), 29–38. https://doi.org/10.4062/biomolther.2017.179.
Lubos, E., Loscalzo, J., & Handy, D. E. (2011). Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxidants & Redox Signaling, 15(7), 1957–1997. https://doi.org/10.1089/ars.2010.3586.
Nita, M., & Grzybowski, A. (2016). The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxidative Medicine and Cellular Longevity, 2016, 3164734. https://doi.org/10.1155/2016/3164734.
Read, A. D., Bentley, R. E., Archer, S. L., & Dunham-Snary, K. J. (2021). Mitochondrial iron-sulfur clusters: Structure, function, and an emerging role in vascular biology. Redox Biology, 47, 102164. https://doi.org/10.1016/j.redox.2021.102164.
Fukai, T., & Ushio-Fukai, M. (2011). Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxidants & Redox Signaling, 15(6), 1583–1606. https://doi.org/10.1089/ars.2011.3999.
Ren, X., Zou, L., Zhang, X., Branco, V., Wang, J., Carvalho, C., Holmgren, A., & Lu, J. (2017). Redox signaling mediated by thioredoxin and glutathione systems in the central nervous system. Antioxidants & Redox Signaling, 27(13), 989–1010. https://doi.org/10.1089/ars.2016.6925.
Schieber, M., & Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. Current Biology: CB, 24(10), R453–R462. https://doi.org/10.1016/j.cub.2014.03.034.
Zhao, Z. (2019). Iron and oxidizing species in oxidative stress and Alzheimer’s disease. Aging Medicine (Milton (N.S.W)), 2(2), 82–87. https://doi.org/10.1002/agm2.12074.
Hill, J. O., Wyatt, H. R., & Peters, J. C. (2012). Energy balance and obesity. Circulation, 126(1), 126–132. https://doi.org/10.1161/CIRCULATIONAHA.111.087213.
Czech, M. P. (2017). Insulin action and resistance in obesity and type 2 diabetes. Nature Medicine, 23(7), 804–814. https://doi.org/10.1038/nm.4350.
Dobrzyn, P., Dobrzyn, A., Miyazaki, M., Cohen, P., Asilmaz, E., Hardie, D. G., Friedman, J. M., & Ntambi, J. M. (2004). Stearoyl-CoA desaturase 1 deficiency increases fatty acid oxidation by activating AMP-activated protein kinase in liver. Proceedings of the National Academy of Sciences of the United States of America, 101(17), 6409–6414. https://doi.org/10.1073/pnas.0401627101.
Larsen, P. J., Fledelius, C., Knudsen, L. B., & Tang-Christensen, M. (2001). Systemic administration of the long-acting GLP-1 derivative NN2211 induces lasting and reversible weight loss in both normal and obese rats. Diabetes, 50(11), 2530–2539. https://doi.org/10.2337/diabetes.50.11.2530.
Farr, O. M., Li, C. R., & Mantzoros, C. S. (2016). Central nervous system regulation of eating: Insights from human brain imaging. Metabolism: Clinical and Experimental, 65(5), 699–713. https://doi.org/10.1016/j.metabol.2016.02.002.
Wu, C., Kang, J. E., Peng, L. J., Li, H., Khan, S. A., Hillard, C. J., Okar, D. A., & Lange, A. J. (2005). Enhancing hepatic glycolysis reduces obesity: Differential effects on lipogenesis depend on site of glycolytic modulation. Cell Metabolism, 2(2), 131–140. https://doi.org/10.1016/j.cmet.2005.07.003.
Foretz, M., Even, P. C., & Viollet, B. (2018). AMPK activation reduces hepatic lipid content by increasing fat oxidation in vivo. International Journal of Molecular Sciences, 19(9), 2826. https://doi.org/10.3390/ijms19092826.
Liangpunsakul, S., Ross, R. A., & Crabb, D. W. (2013). Activation of carbohydrate response element-binding protein by ethanol. Journal of Investigative Medicine: The Official Publication of the American Federation for Clinical Research, 61(2), 270–277. https://doi.org/10.2310/JIM.0b013e31827c2795.
Iizuka, K., Takao, K., & Yabe, D. (2020). ChREBP-mediated regulation of lipid metabolism: Involvement of the gut microbiota, liver, and adipose tissue. Frontiers in Endocrinology, 11, 587189. https://doi.org/10.3389/fendo.2020.587189.
Li, Y., Xu, S., Mihaylova, M. M., Zheng, B., Hou, X., Jiang, B., Park, O., Luo, Z., Lefai, E., Shyy, J. Y., Gao, B., Wierzbicki, M., Verbeuren, T. J., Shaw, R. J., Cohen, R. A., & Zang, M. (2011). AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metabolism, 13(4), 376–388. https://doi.org/10.1016/j.cmet.2011.03.009.
Guo, K., Tian, Q., Yang, L., & Zhou, Z. (2021). The role of glucagon in glycemic variability in type 1 diabetes: A narrative review. Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy, 14, 4865–4873. https://doi.org/10.2147/DMSO.S343514.
Jiang, S., Young, J. L., Wang, K., Qian, Y., & Cai, L. (2020). Diabetic‑induced alterations in hepatic glucose and lipid metabolism: The role of type 1 and type 2 diabetes mellitus (Review). Molecular medicine reports, 22(2), 603–611. https://doi.org/10.3892/mmr.2020.11175.
Sharabi, K., Tavares, C. D., Rines, A. K., & Puigserver, P. (2015). Molecular pathophysiology of hepatic glucose production. Molecular Aspects of Medicine, 46, 21–33. https://doi.org/10.1016/j.mam.2015.09.003.
Zhang, D., Wei, Y., Huang, Q., Chen, Y., Zeng, K., Yang, W., Chen, J., & Chen, J. (2022). Important hormones regulating lipid metabolism. Molecules (Basel, Switzerland), 27(20), 7052. https://doi.org/10.3390/molecules27207052.
Henly, D. C., Phillips, J. W., & Berry, M. N. (1996). Suppression of glycolysis is associated with an increase in glucose cycling in hepatocytes from diabetic rats. The Journal of Biological Chemistry, 271(19), 11268–11271. https://doi.org/10.1074/jbc.271.19.11268.
Giacco, F., & Brownlee, M. (2010). Oxidative stress and diabetic complications. Circulation Research, 107(9), 1058–1070. https://doi.org/10.1161/CIRCRESAHA.110.223545.
Li, Y. G., Ji, D. F., Zhong, S., Lv, Z. Q., & Lin, T. B. (2013). Cooperative anti-diabetic effects of deoxynojirimycin-polysaccharide by inhibiting glucose absorption and modulating glucose metabolism in streptozotocin-induced diabetic mice. PloS One, 8(6), e65892. https://doi.org/10.1371/journal.pone.0065892.
Bence, K. K., & Birnbaum, M. J. (2021). Metabolic drivers of non-alcoholic fatty liver disease. Molecular Metabolism, 50, 101143. https://doi.org/10.1016/j.molmet.2020.101143.
Onyango, A. N. (2022). Excessive gluconeogenesis causes the hepatic insulin resistance paradox and its sequelae. Heliyon, 8(12), e12294. https://doi.org/10.1016/j.heliyon.2022.e12294.
Go, Y., Jeong, J. Y., Jeoung, N. H., Jeon, J. H., Park, B. Y., Kang, H. J., Ha, C. M., Choi, Y. K., Lee, S. J., Ham, H. J., Kim, B. G., Park, K. G., Park, S. Y., Lee, C. H., Choi, C. S., Park, T. S., Lee, W. N., Harris, R. A., & Lee, I. K. (2016). Inhibition of pyruvate dehydrogenase kinase 2 protects against hepatic steatosis through modulation of tricarboxylic acid cycle anaplerosis and ketogenesis. Diabetes, 65(10), 2876–2887. https://doi.org/10.2337/db16-0223.
Ahmad, W., Ijaz, B., Shabbiri, K., Ahmed, F., & Rehman, S. (2017). Oxidative toxicity in diabetes and Alzheimer’s disease: Mechanisms behind ROS/ RNS generation. Journal of Biomedical Science, 24(1), 76. https://doi.org/10.1186/s12929-017-0379-z.
Talwar, D., Miller, C. G., Grossmann, J., Szyrwiel, L., Schwecke, T., Demichev, V., Mikecin Drazic, A. M., Mayakonda, A., Lutsik, P., Veith, C., Milsom, M. D., Müller-Decker, K., Mülleder, M., Ralser, M., & Dick, T. P. (2023). The GAPDH redox switch safeguards reductive capacity and enables survival of stressed tumour cells. Nature Metabolism, 5(4), 660–676. https://doi.org/10.1038/s42255-023-00781-3.
Jellinger, K. A. (2010). Basic mechanisms of neurodegeneration: A critical update. Journal of Cellular and Molecular Medicine, 14(3), 457–487. https://doi.org/10.1111/j.1582-4934.2010.01010.x.
Bouter, C., Henniges, P., Franke, T. N., Irwin, C., Sahlmann, C. O., Sichler, M. E., Beindorff, N., Bayer, T. A., & Bouter, Y. (2019). 18F-FDG-PET detects drastic changes in brain metabolism in the Tg4-42 model of Alzheimer’s disease. Frontiers in Aging Neuroscience, 10, 425. https://doi.org/10.3389/fnagi.2018.00425.
Murphy, M. P., & LeVine, 3rd, H. (2010). Alzheimer’s disease and the amyloid-beta peptide. Journal of Alzheimer’s Disease: JAD, 19(1), 311–323. https://doi.org/10.3233/JAD-2010-1221.
Lau, A., Beheshti, I., Modirrousta, M., Kolesar, T. A., Goertzen, A. L., & Ko, J. H. (2021). Alzheimer’s disease-related metabolic pattern in diverse forms of neurodegenerative diseases. Diagnostics (Basel, Switzerland), 11(11), 2023. https://doi.org/10.3390/diagnostics11112023.
Duffy, P. E., Rapport, M., & Graf, L. (1980). Glial fibrillary acidic protein and Alzheimer-type senile dementia. Neurology, 30(7 Pt 1), 778–782. https://doi.org/10.1212/wnl.30.7.778.
Butterfield, D. A., Favia, M., Spera, I., Campanella, A., Lanza, M., & Castegna, A. (2022). Metabolic features of brain function with relevance to clinical features of Alzheimer and Parkinson diseases. Molecules (Basel, Switzerland), 27(3), 951. https://doi.org/10.3390/molecules27030951.
Demetrius, L. A., Magistretti, P. J., & Pellerin, L. (2015). Alzheimer’s disease: The amyloid hypothesis and the Inverse Warburg effect. Frontiers in Physiology, 5, 522. https://doi.org/10.3389/fphys.2014.00522.
Zhang, X., Alshakhshir, N., & Zhao, L. (2021). Glycolytic metabolism, brain resilience, and Alzheimer’s disease. Frontiers in Neuroscience, 15, 662242. https://doi.org/10.3389/fnins.2021.662242.
Bell, S. M., Burgess, T., Lee, J., Blackburn, D. J., Allen, S. P., & Mortiboys, H. (2020). Peripheral glycolysis in neurodegenerative diseases. International Journal of Molecular Sciences, 21(23), 8924. https://doi.org/10.3390/ijms21238924.
Kaminsky, Y. G., Reddy, V. P., Ashraf, G. M., Ahmad, A., Benberin, V. V., Kosenko, E. A., & Aliev, G. (2013). Age-related defects in erythrocyte 2,3-diphosphoglycerate metabolism in dementia. Aging and Disease, 4(5), 244–255. https://doi.org/10.14336/AD.2013.0400244.
Griffin, J. W., & Bradshaw, P. C. (2017). Amino acid catabolism in Alzheimer’s disease brain: Friend or foe? Oxidative Medicine and Cellular Longevity, 2017, 5472792. https://doi.org/10.1155/2017/5472792.
Cai, R., Zhang, Y., Simmering, J. E., Schultz, J. L., Li, Y., Fernandez-Carasa, I., Consiglio, A., Raya, A., Polgreen, P. M., Narayanan, N. S., Yuan, Y., Chen, Z., Su, W., Han, Y., Zhao, C., Gao, L., Ji, X., Welsh, M. J., & Liu, L. (2019). Enhancing glycolysis attenuates Parkinson’s disease progression in models and clinical databases. The Journal of Clinical Investigation, 129(10), 4539–4549. https://doi.org/10.1172/JCI129987.
Mazzio, E. A., Reams, R. R., & Soliman, K. F. (2004). The role of oxidative stress, impaired glycolysis and mitochondrial respiratory redox failure in the cytotoxic effects of 6-hydroxydopamine in vitro. Brain Research, 1004(1-2), 29–44. https://doi.org/10.1016/j.brainres.2003.12.034.
Tian, Q., Tang, H. L., Tang, Y. Y., Zhang, P., Kang, X., Zou, W., & Tang, X. Q. (2022). Hydrogen sulfide attenuates the cognitive dysfunction in Parkinson’s disease rats via promoting hippocampal microglia M2 polarization by enhancement of hippocampal Warburg effect. Oxidative Medicine and Cellular Longevity, 2022, 2792348. https://doi.org/10.1155/2022/2792348.
Guo, C., Sun, L., Chen, X., & Zhang, D. (2013). Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regeneration Research, 8(21), 2003–2014. https://doi.org/10.3969/j.issn.1673-5374.2013.21.009.
Kierans, S. J., & Taylor, C. T. (2021). Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. The Journal of Physiology, 599(1), 23–37. https://doi.org/10.1113/JP280572.
Bonora, M., Patergnani, S., Rimessi, A., De Marchi, E., Suski, J. M., Bononi, A., Giorgi, C., Marchi, S., Missiroli, S., Poletti, F., Wieckowski, M. R., & Pinton, P. (2012). ATP synthesis and storage. Purinergic Signalling, 8(3), 343–357. https://doi.org/10.1007/s11302-012-9305-8.
Abhishek, S., Deeksha, W., Nethravathi, K. R., Davari, M. D., & Rajakumara, E. (2023). Allosteric crosstalk in modular proteins: Function fine-tuning and drug design. Computational and Structural Biotechnology Journal, 21, 5003–5015. https://doi.org/10.1016/j.csbj.2023.10.013.
Wei, X., Hou, Y., Long, M., Jiang, L., & Du, Y. (2022). Molecular mechanisms underlying the role of hypoxia-inducible factor-1 α in metabolic reprogramming in renal fibrosis. Frontiers in Endocrinology, 13, 927329. https://doi.org/10.3389/fendo.2022.927329.
Hoxhaj, G., & Manning, B. D. (2020). The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nature Reviews Cancer, 20(2), 74–88. https://doi.org/10.1038/s41568-019-0216-7.
Hu, C. J., Wang, L. Y., Chodosh, L. A., Keith, B., & Simon, M. C. (2003). Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Molecular and Cellular Biology, 23(24), 9361–9374. https://doi.org/10.1128/MCB.23.24.9361-9374.2003.
He, W., Batty-Stuart, S., Lee, J. E., & Ohh, M. (2021). HIF-1α hydroxyprolines modulate oxygen-dependent protein stability via single VHL interface with comparable effect on ubiquitination rate. Journal of Molecular Biology, 433(22), 167244. https://doi.org/10.1016/j.jmb.2021.167244.
Strowitzki, M. J., Cummins, E. P., & Taylor, C. T. (2019). Protein hydroxylation by hypoxia-inducible factor (HIF) hydroxylases: Unique or ubiquitous? Cells, 8(5), 384. https://doi.org/10.3390/cells8050384.
Lancaster, D. E., McNeill, L. A., McDonough, M. A., Aplin, R. T., Hewitson, K. S., Pugh, C. W., Ratcliffe, P. J., & Schofield, C. J. (2004). Disruption of dimerization and substrate phosphorylation inhibit factor inhibiting hypoxia-inducible factor (FIH) activity. The Biochemical Journal, 383(Pt. 3), 429–437. https://doi.org/10.1042/BJ20040735.
Ziello, J. E., Jovin, I. S., & Huang, Y. (2007). Hypoxia-Inducible Factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale Journal of Biology and Medicine, 80(2), 51–60.
Yfantis, A., Mylonis, I., Chachami, G., Nikolaidis, M., Amoutzias, G. D., Paraskeva, E., & Simos, G. (2023). Transcriptional response to hypoxia: The role of HIF-1-associated co-regulators. Cells, 12(5), 798. https://doi.org/10.3390/cells12050798.
Nagao, A., Kobayashi, M., Koyasu, S., Chow, C. C. T., & Harada, H. (2019). HIF-1-dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. International Journal of Molecular Sciences, 20(2), 238. https://doi.org/10.3390/ijms20020238.
Gao, S., Zhou, J., Zhao, Y., Toselli, P., & Li, W. (2013). Hypoxia-response element (HRE)-directed transcriptional regulation of the rat lysyl oxidase gene in response to cobalt and cadmium. Toxicological Sciences: An Official Journal of the Society of Toxicology, 132(2), 379–389. https://doi.org/10.1093/toxsci/kfs327.
Kim, J. W., Tchernyshyov, I., Semenza, G. L., & Dang, C. V. (2006). HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metabolism, 3(3), 177–185. https://doi.org/10.1016/j.cmet.2006.02.002.
Cui, X. G., Han, Z. T., He, S. H., Wu, X. D., Chen, T. R., Shao, C. H., Chen, D. L., Su, N., Chen, Y. M., Wang, T., Wang, J., Song, D. W., Yan, W. J., Yang, X. H., Liu, T., Wei, H. F., & Xiao, J. (2017). HIF1/2α mediates hypoxia-induced LDHA expression in human pancreatic cancer cells. Oncotarget, 8(15), 24840–24852. https://doi.org/10.18632/oncotarget.15266.
Pérez-Escuredo, J., Van Hée, V. F., Sboarina, M., Falces, J., Payen, V. L., Pellerin, L., & Sonveaux, P. (2016). Monocarboxylate transporters in the brain and in cancer. Biochimica et Biophysica Acta, 1863(10), 2481–2497. https://doi.org/10.1016/j.bbamcr.2016.03.013.
Zhang, H., Bosch-Marce, M., Shimoda, L. A., Tan, Y. S., Baek, J. H., Wesley, J. B., Gonzalez, F. J., & Semenza, G. L. (2008). Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. The Journal of Biological Chemistry, 283(16), 10892–10903. https://doi.org/10.1074/jbc.M800102200.
Hirota, K. (2021). HIF-α prolyl hydroxylase inhibitors and their implications for biomedicine: A comprehensive review. Biomedicines, 9(5), 468. https://doi.org/10.3390/biomedicines9050468.
Acknowledgements
We would like to thank the Council of Scientific and Industrial Research [CSIR File no-09/028(1112)/2019-EMR-I] for funding AM’s fellowship.
Author information
Authors and Affiliations
Contributions
Conceptualization of the project was done by S.G., R.S. and A.M. A.M. prepared the original draft, which was edited by S.G. and R.S. S.G. and R.S. are corresponding authors of this manuscript. All approve the final version of the submitted manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Malla, A., Gupta, S. & Sur, R. Glycolytic enzymes in non-glycolytic web: functional analysis of the key players. Cell Biochem Biophys 82, 351–378 (2024). https://doi.org/10.1007/s12013-023-01213-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-023-01213-5