Abstract
Coronavirus disease-19 (COVID-19) is caused by the severe acute Respiratory syndrome coronavirus-2 (SARS-CoV-2), which has become unstoppable, spreading rapidly worldwide and, consequently, reaching a pandemic level. This review aims to provide the information available so far on the likely animal origin of SARS-CoV-2 and its possible hosts/reservoirs as well as all natural animal infections and experimental evidence using animal models. Horseshoe bats from the species Rhinolophus affinis seem to be a natural reservoir and pangolins (Manis javanica) appear to be an intermediate host of SARS-CoV-2. Humans remain the most likely spreading source of SARS-CoV-2 to other humans and also to domestic, zoo and farm animals. Indeed, human-to-animal transmission has been reported in cats, dogs, tigers, lions, a puma and minks. Animal-to-human transmission is not a sustained pathway, although mink-to-human transmission remains to be elucidated. Through experimental infections, other animals seem also to be susceptible hosts for SARS-CoV-2, namely ferrets, some non-human primate species, hamsters and transgenic mice, while dogs, pigs and poultry are resistant. A One Health perspective must be implemented in order to develop epidemiological surveillance and establish disease control mechanisms to limit zoonotic transmission. Moreover, research in this field is important to better understand SARS-CoV-2 and to obtain the long-awaited vaccine and specific treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In December 2019, 27 cases of pneumonia of unknown aetiology were identified in Wuhan City, Hubei Province, China (Abdel-Moneim and Abdelwhab 2020; Sohrabi et al. 2020; Zhu et al. 2020). These patients were epidemiologically linked to Wuhan’s Huanan Seafood Wholesale Market, where they were exposed to wildlife animals (Sohrabi et al. 2020; Ji et al. 2020; Rothan and Byrareddy 2020).
At present, we know this is the third outbreak with a highly virulent and large-scale pandemic coronavirus (CoV) causing severe pneumonia in humans in the twenty-first century (Guo et al. 2020; Yang et al. 2020; Wong et al. 2020). The first outbreak, caused by Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), had origin in Guangdong Province, China, beginning in late 2002 and lasting until 2004 (Wong et al. 2020). Middle East Respiratory Syndrome Coronavirus (MERS-CoV) was responsible for the second outbreak and was isolated for the first time from a male patient who was hospitalized with acute pneumonia in Saudi Arabia in 2012 (Wong et al. 2020; Ludwig and Zarbock 2020). Middle East Respiratory Syndrome (MERS) is still an ongoing zoonotic disease largely centered on the Arabian Peninsula (Ludwig and Zarbock 2020; Zhang and Holmes 2020). The World Health Organization (WHO) has named the current pandemic disease as coronavirus disease 2019 (COVID-19) (Sohrabi et al. 2020; Ferrari et al. 2020) and its aetiological agent was designated as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by the Coronavirus Study Group (CSG) of the International Committee on Taxonomy of Viruses (ICTV) (Gorbalenya et al. 2020; Jiang et al. 2020). The WHO has also confirmed the association between this viral pandemic and that wet market in Wuhan City. However, no specific animal origin has been identified so far (Adhikari et al. 2020). SARS-CoV-2 has become unstoppable, spreading rapidly worldwide and consequently reaching a pandemic level since this novel CoV has infected more than 100,000 people in 100 countries worldwide (Ahn et al. 2020; Remuzzi and Remuzzi 2020). At the time of writing (December 3rd, 2020), 64,570,755 people have been infected in 191 countries and 1,494,304 patients have died (Johns Hopkins Coronavirus Resource Center 2020).
As it is a recent disease, there is still much information about the virus that is unknown, so finding an effective antiviral therapy and developing a vaccine are extremely challenging tasks (Zhai et al. 2020). Although successful animal-to-human transitions are rare events, coronaviruses (CoV) have a wide distribution in animals, a high genetic diversity and frequent recombination of their genomes (Ludwig and Zarbock 2020; Voskarides 2020; Wang et al. 2020a). Moreover, CoV seem to transit relatively easily from animals to humans and our globalized world favour such occurrence (Voskarides 2020). Thereby, it is imperative to identify the animal source of SARS-CoV-2 and interspecies movement in order to implement specific control measures and predict and prevent future pandemics, since novel CoV are likely to emerge in humans (Wang et al. 2020a; Anand et al. 2020; Ye et al. 2020).
This paper intended to provide the information available so far regarding the likely animal origin of SARS-CoV-2, its possible hosts/reservoirs as well as all natural animal infections and experimental evidence using animal models.
Potential animal origin and natural reservoir of SARS-CoV-2
At the early stages of the outbreak, there was speculation that SARS-CoV-2 could be a laboratory constructed or manipulated virus (Rabi et al. 2020). However, Andersen et al. (Andersen et al. 2020) concluded that the binding of SARS-CoV-2 is not optimal based on computational analysis, although SARS-CoV-2 has an optimized polybasic (furin) cleavage site in the spike protein and a receptor-binding domain (RBD) with high affinity to angiotensin-converting enzyme 2 (ACE2) from humans, ferrets, cats and other species (Andersen et al. 2020; Hoffmann et al. 2020). These facts suggest that there is another mechanism of binding resulting from the natural selection of the virus in the human or human like ACE2 (Anand et al. 2020; Rabi et al. 2020).
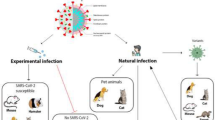
All CoV that have caused disease to humans were of animal origin, especially from bats, which are undoubtedly an important natural reservoir, since the viruses are well adapted and are not pathogenic to them, although they reveal great genetic diversity (Ludwig and Zarbock 2020; Ye et al. 2020; Rabi et al. 2020). Moreover, bat SARS-like CoV exhibit frequent recombination within viral structural proteins between CoV from different hosts, which may increase the potential for cross-species transmission (Ji et al. 2020). Lu et al. (Lu et al. 2020a) analyzed the genome from nine patients (eight of whom had visited the Huanan seafood market in Wuhan) and showed that SARS-CoV-2 was more related to two SARS-like bat CoV from Zhoushan in eastern China: bat-SL-CoVZC45 (with 87.99% identity) and bat-SL-CoVZXC21 (with 87.23% identity), and more distant from SARS-CoV (about 79%) and MERS-CoV (50%) (Wang et al. 2020a; Anand et al. 2020; Lu et al. 2020a). At the protein level, there were only minor insertions or deletions on the proteins encoded by SARS-CoV-2 and these two SARS-like bat CoV (Lu et al. 2020a). However, their study also revealed that the receptor binding protein spike (S) gene of SARS-CoV-2 had only 75% of sequence identity with bat-SL-bat-SL-CoVZC45 and bat-SL-CoVZXC21 (Anand et al. 2020; Lu et al. 2020a), being closer to that of SARS-CoV, despite some variations in the key residues existing at amino acid level (Wang et al. 2020a; Lu et al. 2020a; Xu et al. 2020a). Zhou et al. (Zhou et al. 2020) pointed out that SARS-CoV-2 has 96.2% overall genetic similarity to another betacoronavirus: a horseshoe bat SARSr-CoV designated RaTG13 (Zhou et al. 2020; Shi et al. 2020). In addition, SARS-CoV-2 seems to be a mutated version of bat CoV-RaTG13, detected and isolated in bats from the species Rhinolophus affinis from Yunnan Province, between 2015 and 2017 (Andersen et al. 2020; Zhou et al. 2020). This suggests that bat SARS-like CoVs and human SARS-CoV-2 might share the same ancestor (Guo et al. 2020). Moreover, an insertion of “PAA” on the cleavage site between S1 and S2 was found in a CoV genome identified in bats belonging to the species Rhinolophus malaynus (Zhou et al. 2020). These results suggest that bats are likely the animal reservoir of SARS-CoV-2 (Wong et al. 2020; Zhou et al. 2020) (Fig. 1), but not its primary host, since bat CoV-RaTG13 seems not use the same ACE2 receptor used by SARS-CoV-2 due to its sequence divergence in the RBD that shares only 89% identity in amino acid sequence with that of SARS-CoV-2 (Yuen et al. 2020). Furthermore, bats are not available for sale in Huanan Seafood Market (Guo et al. 2020), and most bat species in Wuhan are hibernating in December (Sun et al. 2020). Moreover, there are no confirmation about direct transmission of SARS-CoV-2 from bats to humans (Wong et al. 2020) and, in fact, the first documented patient was not linked to this market (Huang et al. 2020). Then, despite the role played by that marketplace in the early spreading of SARS-CoV-2 (Adhikari et al. 2020) and its very likely origin in bats, the existence of other sources of infection is assumed (Guo et al. 2020; Huang et al. 2020; Jin et al. 2020; Xiao et al. 2020).
Susceptibility of a range of animals to natural infection by SARS-CoV-2. Bat: Rhinolophus affinis; Turtles: Chrysemys picta bellii, Chelonia mydas and Pelodiscus sinensis; Pangolins: Manis javanica; Snakes: Bungarus multicinctus and Naja atra; Wild felids: Panthera tigris jacksoni, Panthera leo, Puma concolor; Minks: Neovison vison; Cat: Felis catus; Dog: Canis lupus familiaris; Guinea pig: Cavia porcellus; Rabbit: Oryctolagus cuniculus
Potential intermediate hosts of SARS-CoV-2
Given the difficulty of bat-origin viruses being transmitted directly to humans, they need an intermediate host to spread to humans and cause disease (Zhai et al. 2020). In the cases of Severe Acute Respiratory Syndrome (SARS) and MERS, the pathogen was transmitted to humans through exposure to Himalayan palm civet cats (Paguma larvata) and dromedary camels (Camelus dromedarius), respectively (Ludwig and Zarbock 2020; Ahn et al. 2020; Rabi et al. 2020).
It is strongly suggested that, as for SARS-CoV and MERS-CoV, SARS-CoV-2 originated from bats and has been transmitted to other animal hosts and ultimately to humans (Ludwig and Zarbock 2020; Shi et al. 2020; Contini et al. 2020). Presumably, the intermediate hosts of SARS-CoV-2 should be among the wildlife species killed and sold in Wuhan’s Huanan Seafood Wholesale Market (Ye et al. 2020; Huang et al. 2020).
Pangolins
There are studies reporting that pangolins (Manis javanica) appear to be an intermediate host of SARS-CoV-2 (Rabi et al. 2020; Xiao et al. 2020; Boni et al. 2020; Lam et al. 2020; Liu et al. 2020a) (Fig. 1). Malayan pangolins are nocturnal insect-eating mammals found in Southeast Asia but not in China, where the outbreak was first reported (Wong et al. 2020; Zhou et al. 2020). These endangered small animals are illegally smuggled in live from their natural habitats to China by wildlife traffickers who see this trade as very lucrative and on the rise (Wong et al. 2020; Volpato et al. 2020). Pangolins are highly sought after for traditional Chinese medicine, especially their dried scales, in addition to their meat being prized an exclusive delicacy (Volpato et al. 2020).
A group of β-CoV found in pangolins share only about 85–92% nucleotide sequence homology with SARS-CoV-2 (Ye et al. 2020; Yuen et al. 2020; Zhang et al. 2020a), being the second one closest relative to SARS-CoV-2 after bat CoV RaTG13 (Zhang et al. 2020a). However, it is speculated that pangolin ACE2 might show better affinity to SARS-CoV-2, since pangolin β-CoVs contain all six key mutations thought to shape binding to the ACE2 receptor (Zhang and Holmes 2020; Lam et al. 2020; Luan et al. 2020a). It was reported that the RBD of S protein from one sub-lineage of the pangolin CoVs shares 97.4% similarity in amino acid sequences to that of SARS-CoV-2 (Zhang and Holmes 2020; Ye et al. 2020; Zhang et al. 2020a). Likewise, other pangolin-CoV isolated from 17 of 25 Malayan pangolins showed 100%, 98.6%, 97.8% and 90.7% amino acid identity with SARS-CoV-2 in the E, M, N and S proteins, respectively. The RBD of the S protein of pangolin-CoV was similar to that of SARS-CoV-2, with only one difference in a noncritical amino acid (Xiao et al. 2020). Through comparative genomic analysis, the authors suggested that the origin of SARS-CoV-2 resulted from recombination between a virus similar to pangolin-CoV with one similar to RaRG13 (Xiao et al. 2020). The study also reported the occurrence of clinical signs and histopathological changes in infected pangolins as well as the reaction of circulating antibodies against pangolin-CoV with the S protein of SARS-CoV-2 (Xiao et al. 2020). A research group of South China Agricultural University analyzed more than 1000 metagenomic samples and concluded that 70% of pangolins are positive for CoVs. They also reported that the CoV isolated from pangolin shared 99% nucleotide sequence homology with the current infect human strain SARS-CoV-2 (Li et al. 2020; Xu et al. 2020b). In contrast, Deng et al. (Deng et al. 2020) did not detect SARS-CoV-2 antibodies in 17 pangolin serum samples. Supporting these results and the theory that SARS-CoV-2 did not come directly from pangolins, Li et al. (Li et al. 2020) reported that pangolins did not have the RRAR motif, which may be involved in the proteolytic cleavage of the spike protein and, consequently, could impact host range and transmissibility.
Snakes
A study from Ji et al. (Ji et al. 2020) suggests snakes as presumed wildlife animal reservoir (Fig. 1), although there is no reports of SARS-CoV-2 isolation or molecular and serological confirmation of infection. This assumption is based on the virus relative synonymous codon usage (RSCU) bias resembling snake compared with other animals. On the other hand, Zhang et al. (Zhang et al. 2020b) emphasized the controversy generated among virologists by this conclusion, due to the scarcity of biological evidence regarding coronavirus with zoonotic potential to infect organisms other than mammals and birds. Moreover, benchmark results from Zhang et al. (Zhang et al. 2020b) showed that snakes from the species Bungarus multicinctus and Naja atra are not the vertebrates with lowest RSCU distances to SARS-CoV-2. Furthermore, Luan et al. (Luan et al. 2020b) asserted that ACE2 of snakes lost the capability to associate with S protein, so they do not consider this reptile a potential intermediate host.
Turtles
Liu et al. (Liu et al. 2020b) suggested that turtles (Chrysemys picta bellii, Chelonia mydas and Pelodiscus sinensis) may act as a potential intermediate host for SARS-CoV-2 (Fig. 1) based on the key amino acids in ACE2 for interacting with SARS-CoV-2. They reported that more than five residues substitutions were observed in turtle receptors. On the other hand, Luan et al. (Luan et al. 2020b) ruled out the possibility of turtles being intermediate hosts. In their opinion, it is unlikely that reptiles (turtles or other ones) to be infected, considering that all known hosts for CoV are homeothermic animals. Moreover, they analyzed the corresponding amino acids in ACE2 from turtles and concluded that this species does not have the ability to bind to S protein RBD of SARS-CoV-2.
Natural infection in animals
Cats and dogs
It is known that SARS-CoV-2 spike is highly likely to bind to feline ACE2 since only four out of a total of 20 contacting residues are different between feline and human ACE2 (Stout et al. 2020). In order to provide the first evidence of SARS-CoV-2 infection in cats, Zhang et al. (Zhang et al. 2020c) investigated the seroprevalence of SARS-CoV-2 in cats from Wuhan, Hubei Province, by an indirect enzyme-linked immunosorbent assay (ELISA) and virus neutralization test (VNT). The results indicated that cats were infected during the COVID-19 outbreak (Fig. 1). Three cats with the highest titre were owned by three confirmed SARS-CoV-2 infected individuals, suggesting the possibility of direct human-to-cat transmission (Zhang et al. 2020c). On the contrary, a study developed by Deng et al. (Deng et al. 2020) reports that no SARS-CoV-2-specific antibodies were detected in cats (66 pet cats and 21 street cats in Wuhan City). According to this study, the possibility of cats as intermediate host for SARS-CoV-2 is excluded, although their susceptibility to SARS-CoV-2 must be tested by experimental infections.
The study developed by Deng et al. (Deng et al. 2020) also included 487 dogs (90 beagles, 147 pet dogs and 250 street dogs), of which 15 pet dogs and 99 street dogs were from Wuhan City. All of them tested serological negative, even the pet dog from confirmed SARS-CoV-2- infected patient and other two dogs which had close contact with that dog (Deng et al. 2020). The same scenario was observed in France, where viral RNA or antibodies were not detected in dogs (Sailleau et al. 2020; Temmam et al. 2020) neither in cats living with confirmed SARS-CoV-2 infected veterinary students (Temmam et al. 2020). The same was observed in northern Spain, where viral RNA was not detected in 12 dogs and seven cats housed with individuals infected with SARS-CoV-2 (Ruiz-Arrondo et al. 2020). Likewise, in northern Italy, mostly in Lombardy, all 817 companion animals used in a large-scale study to assess SARS-CoV-2 infection tested RT-qPCR negative. However, measurable SARS-CoV-2 neutralizing antibodies titres were detected in 13 dogs (3.4%) and six cats (3.9%) (Patterson et al. 2020).
Although it is extremely unlikely that a pet is going to get COVID-19, there are some cases reporting it (Sit et al. 2020; Centers for Disease Control and Prevention (CDC) 2020) (Tables 1 and 2). However, it remains unclear if pets (as other domestic animals and livestock) are capable of spread the virus to humans (Hernández et al. 2020). Therefore, further studies are needed to reach an assertive conclusion on this subject.
Wild felids
On March 27, 2020 at the Wildlife Conservative Society’s Bronx Zoo in New York City, New York, a Malayan tiger (Panthera tigris jacksoni) showed clinical signs of disease which consisted of a dry cough and some wheezing (World Organization for Animal Health (OIE) 2020; Animal and Plant Health Inspection Service (APHIS) 2020; Wang et al. 2020b). A week later, nasal and oropharyngeal swabs and tracheal wash samples were obtained from the tiger (Wang et al. 2020b). By April 3, three additional tigers (one Malayan tiger and two Amur tigers – Panthera tigris altaica) and three lions (Panthera leo) were showing the same clinical signs (Fig. 1). The animals were isolated and no other animal at the zoo showed any signs of respiratory disease (World Organization for Animal Health (OIE) 2020; Animal and Plant Health Inspection Service (APHIS) 2020). All samples from the first affected tiger were positive by SARS-CoV-2 RT-qPCR testing and gene sequencing (World Organization for Animal Health (OIE) 2020; Wang et al. 2020b). On April 15, the same procedures were performed on an exposed lion confirming its infection by SARS-CoV-2 (World Organization for Animal Health (OIE) 2020). All animals were stable and recovering. It is assumed that an asymptomatic zoo employee infected the animals (World Organization for Animal Health (OIE) 2020; Animal and Plant Health Inspection Service (APHIS) 2020). The same scenario involving three Malayan tigers was reported on October 12th, 2020, at a zoo in Knox, Tennessee. Initially, they showed mild coughing, lethargy and inappetence, however all tigers gradually recovered (World Organization for Animal Health (OIE) 2020).
On July 17, 2020 started an outbreak at a zoo in the city of Johannesburg, South Africa, in which a puma (Puma concolor) tested positive for SARS-CoV-2 by RT-qPCR, after contact with an infected handler (Fig. 1). All other animals in contact with the same person tested negative (World Organization for Animal Health (OIE) 2020).
Minks
Minks (Neovison vison - American mink) were the first intensively farm animals to experience COVID-19 outbreaks, appearing to be a very susceptible species to SARS-CoV-2 (Fig. 1). In the Netherlands, minks from two separate farms in Milheeze and in Beek en Donk displayed mild to severe gastrointestinal and respiratory signs in mid-April 2020, which coincided with their mortality increasing (mortality between 1.2 and 2.4%), especially in pregnant females (Enserink 2020; Molenaar et al. 2020; Oreshkova et al. 2020). The presence of viral RNA was determined by E gene RT-qPCR in different samples, including conchae, throat swab, lung and rectal swab, in addition to the liver and intestines, where viral RNA was less frequently detected. All spleen samples were negative for viral RNA. Some members of one of the farmer’s family and workers from both farmers had respiratory disease symptoms compatible with COVID-19 since the beginning of April. Moreover, some workers had previously tested positive to SARS-CoV-2, the symptoms were present in workers before signs were seen in the minks and the viral sequences obtained from mink samples were related to sequences of human-derived isolates. Therefore, it is plausible that the widespread infection on the mink farms is due to human-to-animal transmission. Nonetheless, mink-to-human transmission is on the table for one worker, according to the preliminary sequencing data. A total of 24 cats found in the surroundings of the farms were sampled for SARS-CoV-2. Seven of them were seropositive, but only one cat was positive for viral RNA. However, it was impossible to generate a sequence from the cat because the amounts of viral RNA were very small. At the first air sampling in the barns was detected low virus load, suggesting dust and/or droplets as possible means of mink-to-mink transmission and occupational risk of exposure for the workers (Enserink 2020; Oreshkova et al. 2020).
In October 2020, an update revealed 62 infected mink farms in the Netherlands. Forty-three were located in the province of Noord Brabant, 17 in the province of Limburg and two in Gelderland. On 25 farms, the owners noticed clinical signs compatible with COVID-19. Currently, humans remain the most likely source of spread of SARS-CoV-2 between farms, therefore additional measures were taken to control the transmission (World Organization for Animal Health (OIE) 2020).
The Dutch Government decided to prepare legislation to end mink farming in the Netherlands in March 2021, before the new breeding period. This decision aims to prevent the establishment of a permanent reservoir in mink industry and a greater risk to public health, if infection by SARS-CoV-2 spills into wild mustelids and other species (World Organization for Animal Health (OIE) 2020).
Between April 26th and November 22nd, 2020, 14 COVID-19 outbreaks occurred in commercial mink farms in Utah, an outbreak in a commercial mink farm in Wisconsin and other in Oregon, USA. Minks were confirmed positive for SARS-CoV-2 based upon molecular testing (RT-qPCR and gene sequencing). Clinical signs included respiratory signs and sudden death of a total of 12,330 minks among 145,757 susceptible animals (World Organization for Animal Health (OIE) 2020).
In mid-May, workers from a mink farm in the municipality of Puebla de Valverde, province of Teruel, Spain, tested positive for COVID-19. All animals (19,500 adults and 73,200 offspring) had not shown clinical signs compatible with the disease. After two tests of samples with negative or inconclusive results, on June 22nd, serum samples and oropharyngeal and rectal swabs of 30 live animals as well as lung parenchyma of six dead animals were tested. One of the oropharyngeal swabs was positive to SARS-CoV-2 by RT-qPCR. On July 7th, 90 oropharyngeal and rectal swabs were collected from 30 adult minks and 60 offspring, resulting in 86.67% of the animals positive to SARS-CoV-2 by RT-qPCR. The results obtained confirm the circulation of SARS-CoV-2 among mink farm animals, without deaths or clinical signs compatible with the disease (World Organization for Animal Health (OIE) 2020).
Between June 15th and August 14th, 2020, SARS-CoV-2 infection has been confirmed in four mink farms (total of 36,200 animals) in the municipalities of Hjørring and Frederikshaw, in Denmark. In the three first farms, some workers tested positive for COVID-19, meanwhile the fourth farm was detected by the new surveillance programme established by the Danish government. Moreover, only on the farm where the first outbreak occurred, the animals did not show clinical signs compatible with COVID-19. Due to precautionary principles, the Danish government decided to cull all minks in the first three farms. On July 20th, 2020, a new strategy to address the problem was implemented. The new strategy is based on a One Health perspective with close cooperation of both local and central authorities. All Danish mink farms are obliged to participate in this new national surveillance programme for SARS-CoV-2 in mink. Furthermore, the Danish Veterinary and Food Administration (DVFA) has made SARS-CoV-2 in mink and ferrets in commercial farms notifiable upon suspicion (World Organization for Animal Health (OIE) 2020). As of September 28th, 2020, the number of infected farms has been updated to 27 (an increase of 23 farms since the last update), all of them in the municipalities of Hjørring and Frederikshaw (World Organization for Animal Health (OIE) 2020).
On November 26th, 2020, Lithuania has also reported an infect mink farm in Jonava, Kaunas, registering 324 deaths of 60,000 susceptible animals. Five farm workers were detected positive to COVID-19 (World Organization for Animal Health (OIE) 2020).
As can be seen, all outbreaks of SARS-CoV-2 in mink have occurred in European countries or in the USA, and the results point to a high susceptibility to the virus by these mustelids.
Other animals
Viral RNA was not detected in samples (oropharyngeal and rectal swabs) obtained from a guinea pig (Cavia porcellus) and two rabbits (Oryctolagus cuniculus) housed with humans with confirmed COVID-19 infections in three households in La Rioja (Northern Spain) (Ruiz-Arrondo et al. 2020) (Fig. 1).
Experimental infection in animals
Ferrets
The studies developed by Kim et al. (Kim et al. 2020) and Shi et al. (Shi et al. 2020) indicated that SARS-COV-2 replicates efficiently, showing high virus titers, in the upper respiratory tract (nasal turbinate, soft palate and tonsils) of ferrets (Mustela putorius furo) (Shi et al. 2020; Stout et al. 2020; Kim et al. 2020) (Fig. 2). The fact that in the study conducted by Shi et al. (Shi et al. 2020) there was no replication in the lower respiratory tract and other organs including heart, liver, spleen, kidneys, pancreas, small intestine and brain raises questions about the possible existence of preventive mechanisms (Shi et al. 2020; Stout et al. 2020). Although at lower levels, viral RNA was also detected in rectal swabs from infected ferrets, which confirms the occurrence of viral replication in the digestive tract (Shi et al. 2020). Indeed, also Kim et al. (Kim et al. 2020) were able to detect viral RNA in urine and faecal specimens, in addition to the detection in serum, saliva, nasal washes, nasal turbinate, trachea, lungs, intestine and kidneys. Moreover, a study conducted by the Erasmus Medical Centre and published in a preprint, provided experimental evidence of transmission of SARS-CoV-2 via direct contact and via the air (via respiratory droplets and/or aerosols) between intranasally inoculated ferrets and naïve ferrets (Richard et al. 2020). Other studies have obtained similar results with regard to the detection of viral RNA in the respiratory tract and other organs, as well as in the lesions observed at necropsy (Schlottau et al. 2020). The reason for the susceptibility of ferrets to SARS-CoV-2 remains unclear. It is suggested that this is due to similarities in the architecture of the respiratory tract between ferrets and humans (Stout et al. 2020; Kim et al. 2020). This further supports ferrets as a suitable model animal for COVID-19 related researches.
Susceptibility of a range of animals to experimental infection by SARS-CoV-2. Cattle: Bos taurus; Mice: Mus musculus; Dog: Canis lupus familiaris; Tree shrew: Tupaia belangeris; Pig: Sus scrofa domesticus; Chicken: Gallus Gallus domesticus; Turkey: Meleagris gallopavo; Duck: Anas platyrhynchos domesticus; Geese: Anser cygnoides; Quail: Coturnix japonica; Ferret: Mustela putorius furo; Cat: Felis catus; Raccoon dog: Nyctereutes procyonoides; Rabbit: Oryctolagus cuniculus; Non-human primates: Macaca mulata; Macaca fascicularis and Callithrix jacchus; Hamster: Mesocricetus auratus
Non-human primates
Luan et al. (Luan et al. 2020b) found that several ACE2 proteins from Primates, Bovidae, Cricetidae and Cetacea maintained the majority of key residues in ACE2 for binding to SARS-CoV-2 RBD. Moreover, through structure simulation of ACE2-RBD complex, the authors concluded that Bovidae and Cricetidae should be included in the screening of intermediate hosts for SARS-CoV-2 since they observed that ACE2 proteins were able to associate with SARS-CoV-2 RBD.
Some articles in press (Bao et al. 2020a; Lu et al. 2020b; Munster et al. 2020; Shan et al. 2020) and published studies (Rockx et al. 2020; Yu et al. 2020) report their results from experimental inoculation of SARS-CoV-2 in non-human primate models. Bao et al. (Bao et al. 2020a) suggested that rhesus monkeys (Macaca mullata) with primary SARS-CoV-2 infection could not be reinfected with the identical strain during their early recovering stage because of humoral immunity stimulated by primary infection.
Some studies support the hypothesis that non-human primates are suitable for preclinical evaluation of anti-viral drugs and vaccines against SARS-CoV-2 since they are permissive to its infection and display COVID-19-like disease (Munster et al. 2020; Rockx et al. 2020; Yu et al. 2020) (Fig. 2). Among them, Gao et al. (Gao et al. 2020) reported that rhesus macaques showed to be a reliable animal model for studying the efficacy of inactivated vaccines against SARS-CoV-2. Lu et al. (Lu et al. 2020b) demonstrated that among the Old World monkeys M. mulatta is the most susceptible to SARS-CoV-2 infection, followed by Macaca fascicularis and Callithrix jacchus. Rockx et al. (Rockx et al. 2020) showed that in relation to M. fascicularis (cynomolgus macaques), SARS-CoV-2 replicates efficiently in upper respiratory tract, which favour the transmission between hosts, and in the lower respiratory tract, resulting in the development of lung disease. According to Yu et al. (Yu et al. 2020), older monkeys are more affected than the younger ones as far as the severity of lung disease is concerned. However, the application of non-human primates for preclinical evaluation is restricted by high costs, availability and the complexity of the necessary management facilities (Di Jiang et al. 2020).
Mice
Mullick et al. (Mullick et al. 2020) emphasized the unique features of SARS-CoV-2 which limit the utility of traditional laboratory animals as mice (Mus musculus), rats (Rattus spp.), rabbits and guinea pigs, since they do not have ACE2 receptors susceptible to SARS-CoV-2 binding. Indeed, regarding mice, some studies have shown that SARS-CoV-2 exhibited limited binding to murine ACE2 (Fig. 2), contrary to the affinity that the virus displays for human ACE2 receptors (hACE2) (Zhou et al. 2020; Lei et al. 2020; Letko et al. 2020; Tai et al. 2020; Wan et al. 2020). Due the low sensitivity of mice to SARS viruses, a murine model – hACE2 transgenic mouse – was used to study the pathogenicity of the virus, along with wild type mice infected with or without SARS-CoV-2 infection (Bao et al. 2020b). Weight loss and virus replication were detected in infected hACE2 mice, as well as lung lesions and pneumonia. However, no important histopathological changes or viral antigens were observed in myocardium, liver, spleen, kidney, brain, intestine and testis. In contrast, any phenomenon was found in wild type mice infected with SARS-CoV-2 (Bao et al. 2020b). Other SARS-CoV-2 hACE2 transgenic mouse – HFH4-hACE2 – was successfully used by Jiang et al. (Di Jiang et al. 2020), and was also used to evaluated the pathogenesis of SARS-CoV and bat SARS-like CoV. The infected mice exhibited typical interstitial pneumonia and pathology. Moreover, significantly weight loss has shown to be closely related to dead or critically ill animals, while animals with less than 20% weight loss have recovered. Thus, weight loss served as a good indicator of disease progression. Viral RNA was predominantly found at the lungs at low viral titre infection, but could also be found in the eye, heart and brain in some mice. Interestingly, HFH4-hACE2 mice that experienced rapid weight loss were found with high viral RNA in the brain. The same scenario occurred when these transgenic mice were infected with bat SARS-like CoV and SARS-CoV in previous studies. This study also revealed that pre-exposure to SARS-CoV-2 could protect mice from reinfection and, consequently, from a potential severe pneumonia. However, it is unclear the role of neutralizing antibody or humoral immunity on the protection from reinfection. Given the results obtained, the authors considered these transgenic mice a useful and valuable animal model for testing vaccines and therapeutics against SARS-CoV-2 infection (Di Jiang et al. 2020). Pruijssers et al. (Pruijssers et al. 2020) applied a therapeutic treatment using remdesivir in infected mice with the chimeric virus. They found a reduction in viral load and an improvement in the clinical condition. According to this study, the antiviral drug remdesivir potently inhibits SARS-CoV-2 in human lung cell cultures, supporting its further clinical testing for treatment of COVID-19. Furthermore, in a recent preprint, remdesivir was used in rhesus macaques and similar results were obtained, which favours the premise that the therapeutic use of this drug should be studied in depth (Williamson et al. 2020).
Hamsters
Some studies established the golden or Syrian hamster (Mesocricetus auratus) as a small model to study the transmission, pathogenesis, treatment and vaccination to SARS-CoV-2, since hamster ACE2 could associate with high affinity to SARS-CoV-2 (Luan et al. 2020b; Chan et al. 2019; Lau et al. 2020) (Fig. 2). In the study conducted by Chan et al. (Chan et al. 2019), primarily inoculated animals developed clinical signs including lethargy, tachypnea and approximately 11% loss of body weight. Viral RNA was detected in the nasal turbinate and trachea. The highest viral titre was observed in the lungs and the lowest levels in the intestine, salivary glands, heart, liver, spleen, lymph nodes, kidney, brain and stool. None of the animals died during the experimental period. However, when euthanized, hamsters revealed pathological changes in the nasal turbinate, trachea and lungs. It was also observed that viral transmission to naïve co-housed hamsters was successful and they showed similar histopathological changes and viral expression in the respiratory tract and extra-pulmonary tissues as the primarily infected hamsters. Nevertheless, in contact hamsters did not suffer reduction in body weight and their passive immunization decreased viral loads in the nasal turbinate and lungs. In other study, male golden hamsters were intranasally inoculated with SARS-CoV-2 virus. Viral RNA was detected with the highest viral load in the lungs and the lowest viral titre in the kidneys and from faecal samples. As in the previous study, viral transmission to naïve co-housed hamsters occurred efficiently. In both groups, hamsters lost more than 10% of the body weight (Sia et al. 2020).
Since SARS-CoV-2 has a considerably more negative impact on elderly, a study was conducted to find if the same scenario occurred in hamsters. This study reported that viral replication in the upper and lower respiratory tract was independent of the age of the animals. However, weight loss was more noticeable in older hamsters and rapid lung recovery was reported only in young hamsters. Moreover, histopathology revealed an early and abundant influx of immune cells in young hamsters (Osterrieder et al. 2020). Similarly, in the study conducted by Boudewijns et al. (Boudewijns et al. 2020), an exuberant innate immune response was identified in golden hamsters, in which signal transducer and activator of transcription 2 (STAT 2) played a dual role, being responsible for severe lung injury but restricting systemic SARS-CoV-2 dissemination.
Raccoon dogs
Raccoon dogs (Nyctereutes procyonoides) seem to be susceptible to SARS-CoV-2 and transmit the virus to contact animals. A study developed by Freuling et al. (Freuling et al. 2020) showed that six out of the nine intranasally inoculated animals developed a productive infection. Effective viral transmission occurred in two out of the three contact animals. The presence of viral RNA and infectious virus in nasal and oropharyngeal swabs were reported in these animals as well as the development of SARS-CoV-2- specific antibody responses. None of the inoculated and contact animals showed clinical signs during the experiment, except mild rhinitis. These results make raccoon dogs a potential intermediate host for SARS-CoV-2 and emphasize the risk that they may pose in transmitting the virus (Freuling et al. 2020).
Cats and dogs
According to Shi et al. (Shi et al. 2020), SARS-CoV-2 can replicate efficiently in cats, especially in younger ones. Indeed, viral RNA was detected in respiratory tissues and the small intestines in cats euthanized at day 3 and day 6, however viral RNA was only detectable in the lungs at day 3. Also, the authors admitted that cats could transmit the virus via respiratory droplets, which makes difficult its control.
Shi et al. (Shi et al. 2020) also found that SARS-CoV-2 replicates poorly in dogs (Fig. 2). They performed oropharyngeal and rectal swabs from five 3-month-old Beagles (which were previously intranasally inoculated with 105 plaque forming unit (PFU) of SARS-CoV-2/CTan/human/2020/Wuhan (CTan-H) and housed with two non-inoculated beagles. Viral RNA was detected in the rectal swabs, but it was not detected infectious virus in any organ or tissue collected from a euthanized dog 4 days post-inoculation. Furthermore, SARS-CoV-2-specific antibodies were detected in two virus-inoculated dogs while the remaining dogs (inoculated and non-inoculated) were seronegative for SARS-CoV-2. These results corroborate the low susceptibility of dogs to SARS-CoV-2.
Tree shrew
The tree shrew, also known as Tupaia belangeris, has been used as an animal model for virus infections (Park et al. 2000; Yang et al. 2013; Li et al. 2018; Sanada et al. 2019; Zhang et al. 2019). Zhao et al. (Zhao et al. 2020) tested this emerging experimental animal susceptibility for SARS-CoV-2 infection. No clinical signs were observed in SARS-CoV-2 inoculated tree shrews, with the exception of increasing body temperature (above 39 °C), particularly in female animals. Limited replication and shedding were detected in infected tree shrews in all three age groups (young, adult and old). Although mild, the main histopathological changes occurred at the pulmonary level. These results confirmed that tree shrew is not susceptible to SARS-CoV-2 infection and, therefore not useful for the study of this new disease (Fig. 2).
Bats
Some studies reported their results about the intranasally inoculation of Egyptian fruit bats (Rousettus aegyptiacus) with SARS-CoV-2 (Schlottau et al. 2020). Bats excreted viruses orally and viral RNA was detected in all bats (co-housed bats too) at higher level in the respiratory tract, but also, in lower levels, in other organs including the heart, skin and intestine. Antibodies were detected in the serum in both inoculated and contact bats (Schlottau et al. 2020). These results imply a potential role of Egyptian fruit bats, which are genetically and immunologically different from horseshoe bats, in replication and transmission of SARS-CoV-2.
Other animals
An experimental infection was performed in three white rabbits (Mykytyn et al. 2020). None of the inoculated animals exhibited clinical signs, nevertheless they developed histopathological signs of moderate inflammation in infected respiratory tissue. These animals showed higher viral RNA shedding in the respiratory tract (nose and throat) than in gastroenteric tract and the development of SARS-CoV-2- specific antibody responses (Mykytyn et al. 2020).
The susceptibility of cattle (Bos taurus) to SARS-CoV-2 was also studied. Two of the six inoculated male Holstein- Friesian calves appeared to be infected since they displayed viral RNA in nasal swabs and specific seroconversion. These results, and taking into account the experimental conditions, showed that SARS-CoV-2 replicate poorly in cattle (Ulrich et al. 2020).
Viral RNA was not detected in any organ samples, contact animals or swabs collected from virus-inoculated pigs (Sus scrofa domesticus), chickens (Gallus Gallus domesticus), ducks (Anas platyrhynchos domesticus), turkeys (Meleagris gallopavo), Japonese quail (Coturnix japonica) and geese (Anser cygnoides). Moreover, no clinical signs were observed and all animals were seronegative for the virus (Fig. 2). Furthermore, SARS-CoV-2 did not replicate in embryonated chicken eggs. Consequently, it is admitted that these species are not susceptible to SARS-CoV-2 (Shi et al. 2020; Schlottau et al. 2020; Suarez et al. 2020).
Summary and concluding remarks
Bats may be the natural reservoir of a SARS-CoV-2-related virus almost identical to SARS-CoV-2. In fact, SARS-CoV-2-related β-CoVs, SARS-CoV-2 and RaTG13 bat virus share the highest genome-wide sequence homology. However, some pangolin SARS-CoV-2-related β-CoVs exhibit strong similarity to SARS-CoV-2 in the RBD, including all six RBD residues. Therefore, it is suggested that pangolins have the potential to act as an intermediate host of SARS-CoV-2.
Live-animal markets promote inter-species contact among wild species, domestic animals and humans. In fact, the epidemiological evidence indicates that the spillover of SARS-CoV-2 to humans was associated with close contact between humans and exotic animals, most likely in Chinese wet markets. A sudden permanent ban of the wild animal trade would promote its shift to the black market. Therefore, stronger action against illegal wildlife trade is imperative as well as strict regulations of exotic animal markets until their complete removal.
To date, humans remain the most likely source of spread of SARS-CoV-2 to other humans, domestic, zoo and farm animals. Actually, the current pandemic is driven by human-to-human transmission. Animal-to-human transmission is not a sustained pathway, although mink-to-human transmission remains to be confirmed. Based on current knowledge, it is unlikely that infected pets play an active role in SARS-CoV-2 transmission to humans. In contrast, several studies and communications showed that companion animals living in areas of high human infection can become infected. Cats are susceptible hosts for the human SARS-CoV-2 likely due to the high degree of similarity between the human and feline forms of ACE2. Further investigation is needed regarding dog susceptibility to SARS-CoV-2. Besides, future research opportunities include wide scale serology surveys of pets in contact with confirmed COVID-19 patients to reveal and evaluate the extension of this transmission route. Other animals are also susceptible hosts for SARS-CoV-2, namely ferrets, probably due to similarities in the architecture of their respective respiratory tracts.
The emergence of other zoonotic infections in the future is inevitable, given the enormous diversity of pathogens, especially in wildlife, and their ongoing evolution. Furthermore, the interaction between humans, animals and the environment can promote their emergence and, consequently, result in infection that, ultimately, can turn into a deadly pandemic. Therefore, is crucial to limit human exposure to animal pathogens as much as possible. Likewise, a G perspective must be implemented in order to develop epidemiological surveillance and establish disease control mechanisms to limit zoonotic transmission through three important means: i) livestock for human consumption; ii) companion animals and iii) exotic animals.
Research studies must be carried out to confirm the origin and natural reservoir of SARS-CoV-2 and to determine the role of other potential reservoirs and animal hosts. Moreover, investigation in this field is important to better understand the pathogenesis of the virus, host-factors as well as continuing to increase knowledge and skills in order to obtain the long-awaited vaccine and specific treatment.
Availability of data and material
Not applicable.
Abbreviations
- ACE2 :
-
Angiotensin converting enzyme 2
- β-CoVs :
-
Betacoronaviruses
- CoV :
-
Coronavirus
- COVID-19 :
-
Coronavirus disease 2019
- CSG :
-
Coronavirus Study Group
- ELISA :
-
Enzyme-linked Immunosorbent Assay
- ICTV :
-
International Committee on Taxonomy of Viruses
- MERS :
-
Middle East respiratory syndrome
- MERS-CoV :
-
Middle East respiratory syndrome coronavirus
- RBD :
-
Receptor-binding domain
- RT-qPCR :
-
Quantitative real-time reverse transcription polymerase chain reaction
- SARS :
-
Severe acute respiratory syndrome
- SARS-CoV :
-
Severe acute respiratory syndrome coronavirus
- SARS-CoV-2 :
-
Severe acute respiratory syndrome coronavirus 2
- STAT 2 :
-
Signal transducer and activator of transcription 2
- VNT :
-
Virus neutralization test
- WHO :
-
World Health Organization
References
Abdel-Moneim AS, Abdelwhab EM (2020) Evidence for SARS-CoV-2 infection of animal hosts. Pathogens 9:1–27. https://doi.org/10.3390/pathogens9070529
Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, Sun C, Sylvia S, Rozelle S, Raat H, Zhou H (2020) Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty 9:1–12. https://doi.org/10.1186/s40249-020-00646-x
Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, Myoung J, Kim BT, Kim SJ (2020) Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). J Microbiol Biotechnol 30:313–324. https://doi.org/10.4014/jmb.2003.03011
Anand KB, Karade S, Sen S, Gupta RM (2020) SARS-CoV-2: Camazotz’s curse. Med J Armed Forces India 76:136–141. https://doi.org/10.1016/j.mjafi.2020.04.008
Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF (2020) The proximal origin of SARS-CoV-2. Nat Med 26:450–452. https://doi.org/10.1038/s41591-020-0820-9
Animal and Plant Health Inspection Service (APHIS) (2020) USDA Statement on the confirmation of COVID-19 in a tiger in New York. US Dep Agric 13:136 https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/NY-zoo-covid-19. Accessed 27 October 2020
Bao L, Deng W, Gao H, Xiao C, Liu J, Xue J, et al. (2020a) Lack of reinfection in rhesus macaques infected with SARS-CoV-2. bioRxiv [Preprint]. https://doi.org/10.1101/2020.03.13.990226
Bao L, Deng W, Huang B, Gao H, Ren L, Wei Q et al (2020b) The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 583:830–833. https://doi.org/10.1038/s41586-020-2312-y
Boni MF, Lemey P, Jiang X, Lam TTY, Perry BW, Castoe TA, Rambaut A, Robertson DL (2020) Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 5:1408–1417. https://doi.org/10.1038/s41564-020-0771-4
Boudewijns R, Thibaut HJ, Kaptein S, Li R, Vergote V, Seldeslachts L, et al. (2020) STAT2 signaling as double-edged sword restricting viral dissemination but driving severe pneumonia in SARS-CoV-2 infected hamsters. bioRxiv [Preprint]. https://doi.org/10.1101/2020.04.23.056838
Centers for Disease Control and Prevention (CDC). Confirmation of COVID-19 in two pet cats in New York. 2020. https://www.cdc.gov/media/releases/2020/s0422-covid-19-cats-NYC.html. Accessed 29 October 2020
Chan JFW, Zhang AJ, Yuan S, Poon VKM, Chan CCS, Lee ACY, Chan WM, Fan Z, Tsoi HW, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai JP, Kok KH, Chu H, Chan KH, Sridhar S, Chen Z, Chen H, To KKW, Yuen KY (2019) Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis 2020:1–50. https://doi.org/10.1093/cid/ciaa325
Contini C, Di Nuzzo M, Barp N, Bonazza A, de Giorgio R, Tognon M et al (2020) The novel zoonotic COVID-19 pandemic: an expected global health concern. J Infect Dev Ctries 14:254–264. https://doi.org/10.3855/jidc.12671
Deng J, Jin Y, Liu Y, Sun J, Hao L, Bai J, Huang T, Lin D, Jin Y, Tian K (2020) Serological survey of SARS-CoV-2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transbound Emerg Dis 67:1745–1749. https://doi.org/10.1111/tbed.13577
Di Jiang R, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR et al (2020) Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 182:50–58. https://doi.org/10.1016/j.cell.2020.05.027
Enserink M (2020) Coronavirus rips through Dutch mink farms, triggering culls. Science. 368:1169. https://doi.org/10.1126/science.368.6496.1169
Ferrari D, Motta A, Strollo M, Banfi G, Locatelli M (2020) Routine blood tests as a potential diagnostic tool for COVID-19. Clin Chem Lab Med 58:1095–1099. https://doi.org/10.1515/cclm-2020-0398
Freuling CM, Breithaupt A, Müller T, Sehl J, Balkema-Bushmann A, Rissmann M, et al. (2020) Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. bioRxiv [Preprint]. https://doi.org/10.1101/2020.08.19.256800
Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, Li Y, Zhu L, Wang N, Lv Z, Gao H, Ge X, Kan B, Hu Y, Liu J, Cai F, Jiang D, Yin Y, Qin C, Li J, Gong X, Lou X, Shi W, Wu D, Zhang H, Zhu L, Deng W, Li Y, Lu J, Li C, Wang X, Yin W, Zhang Y, Qin C (2020) Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 369:77–81. https://doi.org/10.1126/science.abc1932
Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA et al (2020) The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. https://doi.org/10.1038/s41564-020-0695-z
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- an update on the status. Mil Med Res 7:11. https://doi.org/10.1186/s40779-020-00240-0
Hernández M, Abad D, Eiros JM, Rodríguez-Lázaro D (2020) Are animals a neglected transmission route of SARS-CoV-2? Pathogens 9:1–5. https://doi.org/10.3390/pathogens9060480
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:271–280. https://doi.org/10.1016/j.cell.2020.02.052
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. https://doi.org/10.1016/S0140-6736(20)30183-5
Ji W, Wang W, Zhao X, Zai J, Li X (2020) Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol 92:433–440. https://doi.org/10.1002/jmv.25682
Jiang S, Shi Z, Shu Y, Song J, Gao GF, Tan W, Guo D (2020) A distinct name is needed for the new coronavirus. Lancet. 395:949. https://doi.org/10.1016/S0140-6736(20)30419-0
Jin Y, Yang H, Ji W, Wu W, Chen S, Zhang W, Duan G (2020) Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 12:372. https://doi.org/10.3390/v12040372
Johns Hopkins Coronavirus Resource Center. COVID-19 Map. 2020. https://www.coronavirus.jhu.edu/data/mortality%0Ahttps://coronavirus.jhu.edu/map.html. Accessed 3 December 2020
Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MAB, Um J, Song MS, Jeong HW, Lai VD, Kim Y, Chin BS, Park JS, Chung KH, Foo SS, Poo H, Mo IP, Lee OJ, Webby RJ, Jung JU, Choi YK (2020) Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 27:704–709. https://doi.org/10.1016/j.chom.2020.03.023
Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC et al (2020) Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature 583:282–285. https://doi.org/10.1038/s41586-020-2169-0
Lau SY, Wang P, Mok BWY, Zhang AJ, Chu H, Lee ACY, Deng S, Chen P, Chan KH, Song W, Chen Z, To KKW, Chan JFW, Yuen KY, Chen H (2020) Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect 9:837–842. https://doi.org/10.1080/22221751.2020.1756700
Lei C, Qian K, Li T, Zhang S, Fu W, Ding M, Hu S (2020) Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat Commun 11:1–5. https://doi.org/10.1038/s41467-020-16048-4
Letko M, Marzi A, Munster V (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. https://doi.org/10.1038/s41564-020-0688-y
Li R, Yuan B, Xia X, Zhang S, Du Q, Yang C et al (2018) Tree shrew as a new animal model to study the pathogenesis of avian influenza (H9N2) virus infection. Emerg Microbes Infect 7:1–11. https://doi.org/10.1038/s41426-018-0167-1
Li X, Zai J, Zhao Q, Nie Q, Li Y, Foley BT, Chaillon A (2020) Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol 92:602–611. https://doi.org/10.1002/jmv.25731
Liu P, Jiang JZ, Wan XF, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J, Chen J (2020a) Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog 16:595–601. https://doi.org/10.1371/journal.ppat.1008421
Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L (2020b) Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol 92:595–601. https://doi.org/10.1002/jmv.25726
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W (2020a) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 395:565–574. https://doi.org/10.1016/S0140-6736(20)30251-8
Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, et al. (2020b) Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv [Preprint]. https://doi.org/10.1101/2020.04.08.031807
Luan J, Lu Y, Jin X, Zhang L (2020a) Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun 526:165–169. https://doi.org/10.1016/j.bbrc.2020.03.047
Luan J, Jin X, Lu Y, Zhang L (2020b) SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J Med Virol 92:1649–1656. https://doi.org/10.1002/jmv.25817
Ludwig S, Zarbock A (2020) Coronaviruses and SARS-CoV-2: a brief overview. Anesth Analg 131:93–96. https://doi.org/10.1213/ANE.0000000000004845
Molenaar RJ, Vreman S, Hakze-van der Honing RW, Zwart R, de Rond J, Weesendorp E et al (2020) Clinical and pathological findings in SARS-CoV-2 disease outbreaks in farmed mink (Neovison vison). Vet Pathol 57:653–657. https://doi.org/10.1177/0300985820943535
Mullick JB, Simmons CS, Gaire J (2020) Animal models to study emerging technologies against SARS-CoV-2. Cell Mol Bioeng 13:293–303. https://doi.org/10.1007/s12195-020-00638-9
Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER, de Wit E (2020) Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 585:268–272. https://doi.org/10.1038/s41586-020-2324-7
Mykytyn AZ, Lamers MM, Okba NMA, Breugem TI, Schipper D, van den Doel PB, et al. (2020) Susceptibility of rabbits to SARS-CoV-2. bioRxiv [Preprint]. 1–17. https://doi.org/10.1101/2020.08.27.263988
Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Van Der Honing RWH et al (2020) SARS-CoV-2 infection in farmed minks, the Netherlands, April and may 2020. Eurosurveillance. 25:1–7. https://doi.org/10.2807/1560-7917.ES.2020.25.23.2001005
Osterrieder N, Bertzbach LD, Dietert K, Abdelgawad A, Vladimirova D, Kunec D, Hoffmann D, Beer M, Gruber AD, Trimpert J (2020) Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses. 12:779. https://doi.org/10.3390/v12070779
Park US, Su JJ, Ban KC, Qin L, Lee EH, Lee YI (2000) Mutations in the p53 tumor suppressor gene in tree shrew hepatocellular carcinoma associated with hepatitis B virus infection and intake of aflatoxin B1. Gene. 251:73–80. https://doi.org/10.1016/S0378-1119(00)00183-9
Patterson EI, Smith SL, Anderson ER, Patterson GT, Lucente MS, Basano FS, et al. (2020) Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. bioRxiv [Preprint]. https://doi.org/10.1101/2020.07.21.214346
Pruijssers AJ, George AS, Schäfer A, Leist SR, Gralinksi LE, Dinnon KH et al (2020) Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep 32:107940. https://doi.org/10.1016/j.celrep.2020.107940
Rabi FA, Al Zoubi MS, Al-Nasser AD, Kasasbeh GA, Salameh DM (2020) SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 9:1–14. https://doi.org/10.3390/pathogens9030231
Remuzzi A, Remuzzi G (2020) COVID-19 and Italy: what next? Ann Thorac Surg 110:697–700. https://doi.org/10.1016/j.athoracsur.2020.04.003
Richard M, Kok A, de Meulder D, Bestebroer TM, Lamers MM, Okba NMA, Fentener van Vlissingen M, Rockx B, Haagmans BL, Koopmans MPG, Fouchier RAM, Herfst S (2020) SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat Commun 11:3496. https://doi.org/10.1038/s41467-020-17367-2
Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Munnink BBO et al (2020) Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 368:1012–1015. https://doi.org/10.1126/science.abb7314
Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 109:102433. https://doi.org/10.1016/j.jaut.2020.102433
Ruiz-Arrondo I, Portillo A, Palomar AM, Santibáñez S, Santibáñez P, Cervera C et al (2020) Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: a case of an asymptomatic cat with SARS-CoV-2 in Europe. Transbound Emerg Dis 18:10.1111–10.7257. https://doi.org/10.1128/AAC.03728-14
Sailleau C, Dumarest M, Vanhomwegen J, Delaplace M, Caro V, Kwasiborski A, Hourdel V, Chevaillier P, Barbarino A, Comtet L, Pourquier P, Klonjkowski B, Manuguerra JC, Zientara S, le Poder S (2020) First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound Emerg Dis 10(1111):2324–2328. https://doi.org/10.1111/tbed.13659
Sanada T, Yasui F, Honda T, Kayesh MEH, Takano J, Shiogama Y, Yasutomi Y, Tsukiyama-Kohara K, Kohara M (2019) Avian H5N1 influenza virus infection causes severe pneumonia in the northern tree shrew (Tupaia belangeri). Virology. 529:101–110. https://doi.org/10.1016/j.virol.2019.01.015
Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema-Buschmann A, Harder T, Grund C, Hoffmann D, Breithaupt A, Beer M (2020) Experimental transmission studies of SARS-CoV-2 in fruit bats, ferrets, pigs and chickens. SSRN Electron J 1:19–21. https://doi.org/10.2139/ssrn.3578792
Shan C, Yao YF, Yang XL, Zhou YW, Gao G, Peng Y et al (2020) Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in rhesus macaques. Cell Res 30:670–677. https://doi.org/10.1038/s41422-020-0364-z
Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z (2020) Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368:1016–1020. https://doi.org/10.1126/science.abb7015
Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, Peiris M, Yen HL (2020) Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 583:834–838. https://doi.org/10.1038/s41586-020-2342-5
Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW et al (2020) Infection of dogs with SARS-CoV-2. Nature. 586:776–778. https://doi.org/10.1038/s41586-020-2334-5
Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A et al (2020) World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19). Int J Surg 79:163–164. https://doi.org/10.1016/j.ijsu.2020.05.066
Stout AE, André NM, Jaimes JA, Millet JKWG (2020) Coronaviruses in cats and other companion animals: where does SARS-CoV-2/COVID-19 fit? Vet Microbiol 247:108777. https://doi.org/10.1016/j.vetmic.2020.108777
Suarez D, Pantin-Jackwood M, Swayne D, Lee S, DeBlois S, Spackman E (2020) Lack of susceptibility of poultry to SARS-CoV-2 and MERS-CoV. bioRxiv [Preprint]. 2–7. https://doi.org/10.1101/2020.06.16.154658
Sun J, He W, Wang L, Lai A, Ji X, Zhai X et al (2020) COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med 26:483–495. https://doi.org/10.1016/j.molmed.2020.02.008
Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, du L (2020) Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 17:613–620. https://doi.org/10.1038/s41423-020-0400-4
Temmam S, Barbarino A, Maso D, Behillil S, Enouf V, Huon C, Jaraud A, Chevallier L, Backovic M, Pérot P, Verwaerde P, Tiret L, van der Werf S, Eloit M (2020) Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Heal 10:100164. https://doi.org/10.1016/j.onehlt.2020.100164
Ulrich L, Wernike K, Hoffmann D, Mettenleiter TC, Beer M (2020) Experimental infection of cattle with SARS-CoV-2. bioRxiv [Preprint]. https://doi.org/10.1101/2020.08.25.254474
Volpato G, Fontefrancesco MF, Gruppuso P, Zocchi DM, Pieroni A (2020) Baby pangolins on my plate: possible lessons to learn from the COVID-19 pandemic. J Ethnobiol Ethnomed 16:1–12. https://doi.org/10.1186/s13002-020-00366-4
Voskarides K (2020) Animal-to-human viral transitions: is SARS-CoV-2 an evolutionarily successful one? J Mol Evol 88:421–423. https://doi.org/10.1007/s00239-020-09947-z
Wan Y, Shang J, Graham R, Baric RS, Li F (2020) Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94:1–9. https://doi.org/10.1128/jvi.00127-20
Wang H, Li X, Li T, Zhang S, Wang L, Wu X, Liu J (2020a) The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur J Clin Microbiol Infect Dis 39:1629–1635. https://doi.org/10.1007/s10096-020-03899-4
Wang L, Mitchell PK, Calle PP, Bartlett SL, McAloose D, Killian ML, Yuan F, Fang Y, Goodman LB, Fredrickson R, Elvinger F, Terio K, Franzen K, Stuber T, Diel DG, Torchetti MK (2020b) Complete genome sequence of SARS-CoV-2 in a tiger from a U.S. zoological collection. Microbiol Resour Announc 9:e00468–e00420. https://doi.org/10.1128/mra.00468-20
Williamson BN, Feldmann F, Schwarz B, Meade-White K, Porter DP, Schulz J et al (2020) Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 585:273–276. https://doi.org/10.1016/j.lfs.2020.117592
Wong G, Bi YH, Wang QH, Chen XW, Zhang ZG, Yao YG (2020) Zoonotic origins of human coronavirus 2019 (HCoV-19 / SARS-CoV-2): Why is this work important? Zool Res 41:213–219. https://doi.org/10.24272/j.issn.2095-8137.2020.031
World Organization for Animal Health (OIE). World Animal Health Information System. 2020. https://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/WI/index/newlang/en. Accessed 3 December 2020
Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X, Zhang Z, Shu F, Huang W, Li Y, Zhang Z, Chen RA, Wu YJ, Peng SM, Huang M, Xie WJ, Cai QH, Hou FH, Chen W, Xiao L, Shen Y (2020) Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 583:286–289. https://doi.org/10.1038/s41586-020-2313-x
Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong WHP (2020a) Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci 63:457–460. https://doi.org/10.1007/s11427-020-1637-5
Xu J, Zhao S, Teng T, Abdalla AE, Zhu W, Xie L, Wang Y, Guo X (2020b) Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 12:244. https://doi.org/10.3390/v12020244
Yang ZF, Zhao J, Zhu YT, Wang YT, Liu R, Zhao SS, Li RF, Yang CG, Li JQ, Zhong NS (2013) The tree shrew provides a useful alternative model for the study of influenza H1N1 virus. Virol J 10:1–9. https://doi.org/10.1186/1743-422X-10-111
Yang Y, Peng F, Wang R, Guan K, Jiang T, Xu G et al (2020) The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun 109:102434. https://doi.org/10.1016/j.jaut.2020.102434
Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY (2020) Zoonotic origins of human coronaviruses. Int J Biol Sci 16:1686–1697. https://doi.org/10.7150/ijbs.45472
Yu P, Qi F, Xu Y, Li F, Liu P, Liu J, Bao L, Deng W, Gao H, Xiang Z, Xiao C, Lv Q, Gong S, Liu J, Song Z, Qu Y, Xue J, Wei Q, Liu M, Wang G, Wang S, Yu H, Liu X, Huang B, Wang W, Zhao L, Wang H, Ye F, Zhou W, Zhen W, Han J, Wu G, Jin Q, Wang J, Tan W, Qin C (2020) Age-related rhesus macaque models of COVID-19. Anim Model Exp Med 3:93–97. https://doi.org/10.1002/ame2.12108
Yuen KS, Ye ZW, Fung SY, Chan CP, Jin DY (2020) SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci 10:1–5. https://doi.org/10.1186/s13578-020-00404-4
Zhai SL, Wei WK, Lv DH, Xu ZH, Chen QL, Sun MF, Li F, Wang D (2020) Where did SARS-CoV-2 come from? Vet Rec 186:254–25254. https://doi.org/10.1136/vr.m740
Zhang YZ, Holmes EC (2020) A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 181:223–227. https://doi.org/10.1016/j.cell.2020.03.035
Zhang L, Shen ZL, Feng Y, Li DQ, Zhang NN, Deng YQ, Qi XP, Sun XM, Dai JJ, Yang CG, Yang ZF, Qin CF, Xia XS (2019) Infectivity of Zika virus on primary cells support tree shrew as animal model. Emerg Microbes Infect 8:232–241. https://doi.org/10.1080/22221751.2018.1559707
Zhang T, Wu Q, Zhang Z (2020a) Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol 30:1346–1351. https://doi.org/10.1016/j.cub.2020.03.022
Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y (2020b) Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J Proteome Res 19:1351–1360. https://doi.org/10.1021/acs.jproteome.0c00129
Zhang Q, Zhang H, Huang K, Yang Y, Hui X, Gao J et al (2020c) SARS-CoV-2 neutralizing serum antibodies in cats: a serological investigation. Emerg Microbes Infect 9:2013–2019. https://doi.org/10.1101/2020.04.01.021196
Zhao Y, Wang J, Kuang D, Xu J, Yang M, Ma C, Zhao S, Li J, Long H, Ding K, Gao J, Liu J, Wang H, Li H, Yang Y, Yu W, Yang J, Zheng Y, Wu D, Lu S, Liu H, Peng X (2020) Susceptibility of tree shrew to SARS-CoV-2 infection. Sci Rep 10:16007. https://doi.org/10.1038/s41598-020-72563-w
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. https://doi.org/10.1038/s41586-020-2012-7
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. https://doi.org/10.1056/nejmoa2001017
Funding
This study was supported by project UIDB/CVT/00772/2020 funded by Fundação para a Ciência e a Tecnologia (FCT).
Author information
Authors and Affiliations
Contributions
BV performed the literature research, designed the structure of review and wrote the paper. APL, MCF and MS critically reviewed the manuscript. LC and ACC supervised and critically reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
do Vale, B., Lopes, A.P., Fontes, M.d. et al. Bats, pangolins, minks and other animals - villains or victims of SARS-CoV-2?. Vet Res Commun 45, 1–19 (2021). https://doi.org/10.1007/s11259-021-09787-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11259-021-09787-2