Abstract
Background
Male infertility (MI) is a polygenic condition mainly induced by spermatogenic failure/arrest or systemic disease with a large clinical spectrum. Lately, genetic sequencing allowed the identification of several variants implicated in both aforesaid situations.
Methods and results
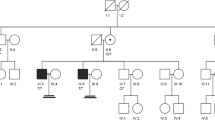
In this case study, we performed whole exome sequencing (WES) on the genomic DNA of a 37-year-old Moroccan man with Non-Obstructive Azoospermia. Results revealed two variants in genes highly expressed in testicular tissue. The first was a heterozygous frameshift variant in the AURKC gene, causing a premature stop codon at position 71 of the AURKC protein, critical for spermatogenesis. The second was a hemizygous missense variant in the ABCD1 gene, resulting in an H299R substitution in the ABCD1 protein, essential for transporting Very Long Chain Fatty Acids (VLCFAs) into peroxisomes. ABCD1 variants are linked to X-linked Adrenoleukodystrophy (X-ALD), a disease caused by VLCFAs accumulation in cells. The patient’s family pedigree suggests X-linked transmission of MI, which may be a subclinical form of late-onset X-ALD in affected members, indicating that the ABCD1 variant likely affects spermatogenesis. This hypothesis is supported by literature linking X-ALD to MI, ABCD1’s high expression in human testes, and the significant impact of the H299R substitution on ABCD1 transporter’s molecular dynamics.
Conclusions
These insights highlight the role of genetic mutations in male infertility, demonstrating that spermatogenesis can be disrupted either directly by specific mutations or indirectly through broader genetic disorders, underscoring the importance of comprehensive genetic testing.






Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Leslie SW, Soon-Sutton TL, Khan MAB (2024) Male infertility. Treasure Island (FL)
Krausz C, Riera-Escamilla A (2018) Genetics of male infertility. Nat Rev Urol 15:369–384. https://doi.org/10.1038/s41585-018-0003-3
O’Neill M, Ruthig VA, Haller M et al (2023) In: Leung PCK, Qiao JBT-HR, Second PG E (eds) Chap. 1 - developmental genetics of the male reproductive system. Academic, pp 3–28
Nenicu A, Lüers GH, Kovacs W et al (2007) Peroxisomes in human and mouse testis: differential expression of peroxisomal proteins in germ cells and distinct somatic cell types of the testis. Biol Reprod 77:1060–1072. https://doi.org/10.1095/biolreprod.107.061242
Cooper GM (2000) Peroxisomes. In: The Cell: A Molecular Approach. 2nd edition
Engelen M, Kemp S, de Visser M et al (2012) X-linked adrenoleukodystrophy (X-ALD): clinical presentation and guidelines for diagnosis, follow-up and management. Orphanet J Rare Dis 7:51. https://doi.org/10.1186/1750-1172-7-51
Cappa M, Todisco T, Bizzarri C (2023) X-linked adrenoleukodystrophy and primary adrenal insufficiency. Front Endocrinol (Lausanne) 14:1309053. https://doi.org/10.3389/fendo.2023.1309053
Aversa A, Palleschi S, Cruccu G et al (1998) Rapid decline of fertility in a case of adrenoleukodystrophy. Hum Reprod 13:2474–2479. https://doi.org/10.1093/humrep/13.9.2474
Suryawanshi A, Middleton T, Ganda K (2015) An unusual presentation of X-linked adrenoleukodystrophy. Endocrinol Diabetes Metab case Rep 2015:150098. https://doi.org/10.1530/EDM-15-0098
Le LTM, Thompson JR, Dang PX et al (2022) Structures of the human peroxisomal fatty acid transporter ABCD1 in a lipid environment. Commun Biol 5:1–8. https://doi.org/10.1038/s42003-021-02970-w
Zhao S, Agafonov O, Azab A et al (2020) Accuracy and efficiency of germline variant calling pipelines for human genome data. Sci Rep 10:20222. https://doi.org/10.1038/s41598-020-77218-4
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med off J Am Coll Med Genet 17:405–424. https://doi.org/10.1038/gim.2015.30
Okonechnikov K, Golosova O, Fursov M, team the U (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Vockel M, Riera-Escamilla A, Tüttelmann F, Krausz C (2021) The X chromosome and male infertility. Hum Genet 140:203–215. https://doi.org/10.1007/s00439-019-02101-w
Redouane S, Charoute H, Harmak H et al (2022) Computational study of the potential impact of AURKC missense SNPs on AURKC-INCENP interaction and their correlation to macrozoospermia. J Biomol Struct Dyn 1–20. https://doi.org/10.1080/07391102.2022.2142846
Fellmeth JE, Ghanaim EM, Schindler K (2016) Characterization of macrozoospermia-associated AURKC mutations in a mammalian meiotic system. Hum Mol Genet 25:2698–2711. https://doi.org/10.1093/hmg/ddw128
Carmena M, Wheelock M, Funabiki H, Earnshaw WC (2012) The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 13:789–803. https://doi.org/10.1038/nrm3474
Ortega V, Oyanedel J, Fleck-Lavergne D et al (2020) Macrozoospermia associated with mutations of AURKC gene: first case report in Latin America and literature review. Rev Int Androl 18:159–163. https://doi.org/10.1016/j.androl.2019.04.004
Höftberger R, Kunze M, Weinhofer I et al (2007) Distribution and cellular localization of adrenoleukodystrophy protein in human tissues: implications for X-linked adrenoleukodystrophy. Neurobiol Dis 28:165–174. https://doi.org/10.1016/j.nbd.2007.07.007
Masters C, Crane D (1996) Chap. 3 The peroxisome. In: Bittar EE, Bittar NBT-P of MB (eds) Cellular Organelles and the Extracellular Matrix. Elsevier, pp 39–58
Eloualid A, Rouba H, Rhaissi H et al (2014) Prevalence of the Aurora kinase C c.144delC mutation in infertile Moroccan men. Fertil Steril 101:1086–1090. https://doi.org/10.1016/j.fertnstert.2013.12.040
van de Stadt SIW, Mooyer PAW, Dijkstra IME et al (2021) Biochemical studies in fibroblasts to interpret variants of unknown significance in the ABCD1 gene. Genes (Basel) 12. https://doi.org/10.3390/genes12121930
Acknowledgements
This research was supported through computational resources of HPC-MARWAN (hpc.marwan.ma) provided by the National Center for Scientific and Technical Research (CNRST), Rabat, Morocco.
Funding
No funds, grants, or other support was received.
Author information
Authors and Affiliations
Contributions
Salaheddine Redouane: Conceptualization, data collection, bioinformatic analysis, writing. Houda Harmak: Review and enhancements of the writing. Adil El Hamouchi and Hicham Charoute: Sequencing and bioinformatic analysis coordination. Noureddine Louanjli: Supervision during the recruitment process of the patient. Abderrahim Malki: Academic supervision and reviewing. Abdelhamid Barakat and Hassan Rouba: Technical supervision and reviewing.
Corresponding author
Ethics declarations
Ethical approval
This study was approved by the Biomedical Research Ethics Committee at the Faculty of Medicine and Pharmacy, Casablanca, Morocco (approval number: 05/2022). We confirm that all research work in this study complies with ethical guidelines, including adherence to the legal requirements of the study country.
Consent to participate
Informed consent was obtained from the recruited participant.
Consent to publish
Patient signed an informed consent form for the publication of his data.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Redouane, S., Harmak, H., El Hamouchi, A. et al. Whole exome sequencing reveals ABCD1 variant as a potential contributor to male infertility. Mol Biol Rep 52, 148 (2025). https://doi.org/10.1007/s11033-025-10234-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-025-10234-7