Abstract
Background
Breast cancer (BC) is the most prevalent cancer among females worldwide. Numerous studies suggest that specific RNAs play a crucial role in carcinogenesis. The primate-specific microRNA gene cluster located on the 19q27.3 region of chromosome 19 (C19MC) could potentially regulate tumor cell proliferation, migration, and invasion.
Objective
The objective of this study was to compare the expression of miRNAs from the C19MC cluster in breast cancer tumor and non-tumor samples, as well as in the serum of individuals affected by BC and healthy individuals.
Methods
Peripheral blood was collected from 100 BC patients and 100 healthy individuals, and breast cancer samples including tumor and margin tissues were obtained. After RNA extraction, Real-time PCR was employed to investigate the expression of C19MC, specifically mir-515–1, mir-515–2, mir-516-A1, mir-516-A2, mir-516-B1, mir-516-B2, mir-517-A, mir-517-B, mir-517-C, and mir-518-A1, in the serum and tissue of BC patients and tumor margins. Statistical analyses and ROC curves were generated using GraphPad Prism software (v8.04), with a significance level set at p < 0.05.
Results
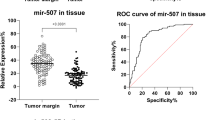
Our findings demonstrate a strong correlation between high expression of all C19MC miRNAs mentioned, except for mir-517-B, mir-517-C and mir- 518 in BC. These miRNAs show potential as notable non-invasive tumor markers.
Conclusion
The data obtained from our study support the overall impact of C19MC miRNAs in BC detection and emphasize the potential role of several C19MC members in this process.


Similar content being viewed by others
Data availability
All data analyzed during this study are included in this published article. Further raw data from the current study are available from the corresponding author upon request.
References
Gopinath A, Cheema AH, Chaludiya K, Khalid M, Nwosu M, Agyeman WY et al (2022) The impact of dietary fat on breast cancer incidence and survival: a systematic review. Cureus 14(10):e30003. https://doi.org/10.7759/cureus.30003. ((PubMed: 36381753))
Kolyvas EA, Caldas C, Kelly K, Ahmad SS (2022) Androgen receptor function and targeted therapeutics across breast cancer subtypes. Breast Cancer Res 24(1):79. https://doi.org/10.1186/s13058-022-01574-4. ((PubMed: 36376977))
Chakrabortty A, Patton DJ, Smith BF, Agarwal P (2023) miRNAs: potential as biomarkers and therapeutic targets for cancer. Genes 14(7):1375. https://doi.org/10.3390/genes14071375
Fridrichova I, Zmetakova I (2019) MicroRNAs contribute to breast cancer invasiveness. Cells 8(11):1361. https://doi.org/10.3390/cells8111361. ((PubMed: 31683635))
Choo KB, Soon YL, Nguyen PNN, Hiew MSY, Huang CJ (2014) MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J Biomed Sci 21:95
Pan G, Liu Y, Shang L, Zhou F, Yang S (2021) EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun (Lond) 41(3):199–217. https://doi.org/10.1002/cac2.12138. ((PubMed: 33506604))
Huang CJ, Nguyen PNN, Choo KB, Sugii S, Wee K, Cheong SK et al (2014) Frequent co-expression of miRNA-5p and -3p species and cross-targeting in induced pluripotent stem cells. Int J Med Sci 11:82
Razak SRA, Ueno K, Takayama N, Nariai N, Nagasaki M, Saito R et al (2013) Profiling of microRNA in human and mouse ES and iPS cells reveals overlapping but distinct microRNA expression patterns. PLoS ONE 8:e73532
Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y et al (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8:376–388
Zhang S, Shan C, Kong G, Du Y, Ye L, Zhang X (2012) MicroRNA-520e suppresses the growth of hepatoma cells by targeting the NF-κB-inducing kinase (NIK). Oncogene 31:3607–3620
Keklikoglou I, Koerner C, Schmidt C, Zhang JD, Heckmann D, Shavinskaya A et al (2012) MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor-negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene 31:4150–4163
Kleinman CL, Gerges N, Papillon-Cavanagh S, Sin-Chan P, Pramatarova A, Quang D-AK et al (2014) Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet 46:39–44
Cui W, Zhang Y, Hu N, Shan C, Zhang S, Zhang W et al (2010) miRNA-520b and mir-520e sensitize breast cancer cells to complement attack via directly targeting 3’UTR of CD46. Cancer Biol Ther 10:232–241
Min D, Lv X, Wang X, Zhang B, Meng W, Yu F et al (2013) Downregulation of mir-302c and mir-520c by 1,25(OH)2D3 treatment enhances the susceptibility of tumor cells to natural killer cell-mediated cytotoxicity. Br J Cancer 109:723–730
Su C-M, Wang M-Y, Hong C-C, Chen H-A, Su Y-H, Wu C-H et al (2016) mir-520h is crucial for DAPK2 regulation and breast cancer progression. Oncogene 35(9):1134–1142
Zhang J, Liu L, Sun Y, Xiang J, Zhou D, Wang L et al (2016) MicroRNA-520g promotes epithelial ovarian cancer progression and chemoresistance via DAPK2 repression. Oncotarget 7:26516–26534
Wang J, Haubrock M, Cao K-M, Hua X, Zhang C-Y, Wingender E et al (2011) Regulatory coordination of clustered microRNAs based on microRNA-transcription factor regulatory network. BMC Syst Biol 5:199
Lin S, Cheung WKC, Chen S, Lu G, Wang Z, Xie D et al (2010) Computational identification and characterization of primate-specific microRNAs in the human genome. Comput Biol Chem 34:232–241
Zhou J-Y, Zheng S-R, Liu J, Shi R, Yu H-L, Wei M (2016) Mir-519d facilitates the progression and metastasis of cervical cancer through direct targeting Smad7. Cancer Cell Int 16:21
Kan H, Guo W, Huang Y, Liu D (2015) MicroRNA-520g induces epithelial-mesenchymal transition and promotes metastasis of hepatocellular carcinoma by targeting SMAD7. FEBS Lett 589:102–109
Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefe A, Coullin P, Moore GE (2010) The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet 19:3566–3582
Sugii S, Kida Y, Berggren WT, Evans RM (2011) Feeder-dependent and feeder-independent iPS cell derivation from human and mouse adipose stem cells. Nat Protoc 6:346–358
Choo KB, Tai L, Hymavathee KS, Wong CY, Nguyen PNN, Huang CJ et al (2014) Oxidative stress-induced premature senescence in Wharton’s jelly-derived mesenchymal stem cells. Int J Med Sci 11:1201–1207
Ma W, Yu Q, Jiang J, Du X, Huang L, Zhao L et al (2016) mir-517a is an independent prognostic marker and contributes to cell migration and invasion in human colorectal cancer. Oncol Lett 11:2583–2589
Ren G, Wang L (2016) Study on the relationship between mir-520g and the development of breast cancer. Eur Rev Med Pharmacol Sci 20:657–663
Tsai C-H, Tsai H-C, Huang H-N, Hung C-H, Hsu C-J, Fong Y-C et al (2014) Resistin promotes tumor metastasis by down-regulation of mir-519d through the AMPK/p38 signaling pathway in human chondrosarcoma cells. Oncotarget 6:258–270
Brownlie RJ, Zamoyska R (2013) T cell receptor signaling networks: branched, diversified and bounded. Nat Rev Immunol 13:257–269
Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA (2015) Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta 1855:104–121
Guan R, Cai S, Sun M, Xu M (2017) Upregulation of miR-520b promotes ovarian cancer growth. Oncol Lett 14:3155–3161. https://doi.org/10.3892/ol.2017.6552
Hu N, Zhang J, Cui W, Kong G, Zhang S, Yue L, Bai X, Zhang Z, Zhang W, Zhang X, Ye L (2011) miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J Biol Chem 286(15):13714–13722
Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R (2022) miRNA: a promising therapeutic target in cancer. Int J Mol Sci 23(19):11502. https://doi.org/10.3390/ijms231911502
Li M, Lee KF, Lu Y, Clarke I, Shih D, Eberhart C et al (2009) Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell 16:533–546
Flor I, Neumann A, Freter C, Helmke BM, Langenbuch M, Rippe V et al (2012) Abundant expression and hemimethylation of C19MC in cell cultures from placenta-derived stromal cells. Biochem Biophys Res Commun 422:411–416
Pinho FG, Frampton AE, Nunes J (2013) Downregulation of microRNA-515-5p by the estrogen receptor modulates sphingosine kinase 1 and breast cancer cell proliferation. Can Res 73:5936–5948
Jagadeeswaran G, Zheng Y, Sumathipala N, Jiang H, Arrese EL, Soulages JL et al (2010) Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics 11:1–18
Li S-C, Liao Y-L, Ho M-R, Tsai K-W, Lai C-H, Lin W (2012) miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics 13(Suppl 1):S13
Carroll AP, Goodall GJ, Liu B (2014) Understanding principles of miRNA target recognition and function through integrated biological and bioinformatics approaches. Wiley Interdisc Rev 5:361–379
Akbari Moqadam F, Pieters R, den Boer ML (2013) The hunting of targets: challenge in miRNA research. Leukemia 27:16–23
Jalvy-Delvaille S, Maurel M, Majo V, Pierre N, Chabas S, Combe C et al (2012) Molecular basis of differential target regulation by mir-96 and mir-182: the glypican-3 as a model. Nucleic Acids Res 40:1356–1365
Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK et al (2008) MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26:2496–2505
Portt L, Norman G, Clapp C, Greenwood M, Greenwood MT (2011) Anti-apoptosis and cell survival: a review. Biochem Biophys Acta 1813:238–259
Acknowledgements
We thank all patients and individuals for participating in this project.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
SARA Experimented and wrote the manuscript with support from MH and MAHF. MH supervised the work and contributed to the design and implementation of the research, the analysis of the results, and the writing of the manuscript. MAHF Helped supervise the project. All authors discussed the results and contributed and reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors confirm that there are no known conflicts of interest associated with this publication, and there has been no significant financial support for this work that could have influencedits outcome.
Ethical approval
This study was approved by the Department of Biology committee at the University of Tabriz (14000907–198-7).
Consent to participate
All stages of sample collection have been done with the consent of patients and healthy people, and written consent forms have been obtained.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Altalebi, S.A.R., Haghi, M. & Hosseinpour Feizi, M.A. Expression study of microRNA cluster on chromosome 19 (C19MC) in tumor tissue and serum of breast cancer patient. Mol Biol Rep 50, 9825–9831 (2023). https://doi.org/10.1007/s11033-023-08801-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08801-x