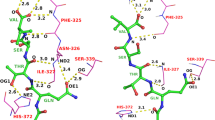
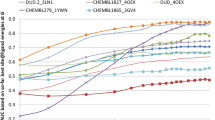
Abstract
The Surflex flexible molecular docking method has been generalized and extended in two primary areas related to the search component of docking. First, incorporation of a small-molecule force-field extends the search into Cartesian coordinates constrained by internal ligand energetics. Whereas previous versions searched only the alignment and acyclic torsional space of the ligand, the new approach supports dynamic ring flexibility and all-atom optimization of docked ligand poses. Second, knowledge of well established molecular interactions between ligand fragments and a target protein can be directly exploited to guide the search process. This offers advantages in some cases over the search strategy where ligand alignment is guided solely by a “protomol” (a pre-computed molecular representation of an idealized ligand). Results are presented on both docking accuracy and screening utility using multiple publicly available benchmark data sets that place Surflex’s performance in the context of other molecular docking methods. In terms of docking accuracy, Surflex-Dock 2.1 performs as well as the best available methods. In the area of screening utility, Surflex’s performance is extremely robust, and it is clearly superior to other methods within the set of cases for which comparative data are available, with roughly double the screening enrichment performance.














Similar content being viewed by others
References
Walters PW, Stahl MT, Murcko MA (1998) Drug Discov Today 3:160–178
Jain AN (1996) J Comput Aided Mol Des 10:427–440
Bohm HJ (1994) J Comput Aided Mol Des 8:243–256
Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP (1997) J Comput Aided Mol Des 11:425–445
Rognan D, Lauemoller SL, Holm A, Buus S, Tschinke V (1999) J Med Chem 42:4650–4658
Wang R, Liu L, Lai L, Tang Y (1998) J Mol Model 4:379–384
Muegge I, Martin YC (1999) J Med Chem 42:791–804
Gohlke H, Hendlich M, Klebe G (2000) J Mol Biol 295:337–356
Welch W, Ruppert J, Jain AN (1996) Chem Biol 3:449–462
Goodsell DS, Morris GM, Olson AJ (1996) J Mol Recognit 9:1–5
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) J Mol Biol 267:727–748
Rarey M, Kramer B, Lengauer T, Klebe G (1996) J Mol Biol 261:470–489
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) J Med Chem 47:1750–1759
Schulz-Gasch T, Stahl M (2003) J Mol Model (Online) 9:47–57
Zavodszky MI, Sanschagrin PC, Korde RS, Kuhn LA (2002) J Comput Aided Mol Des 16:883–902
Jain AN (2003) J Med Chem 46:499–511
Pham TA, Jain AN (2006) J Med Chem 49:5856–5868
Bissantz C, Folkers G, Rognan D (2000) J Med Chem 43:4759–4767
Kellenberger E, Rodrigo J, Muller P, Rognan D (2004) Proteins 57:225–242
Miteva MA, Lee WH, Montes MO, Villoutreix BO (2005) J Med Chem 48:6012–6022
Jain AN (2004) Curr Opin Drug Discov Devel 7:396–403
Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS (2006) J Med Chem 49:5912–5931
Perola E, Walters WP, Charifson PS (2004) Proteins 56:235–249
Cummings MD, DesJarlais RL, Gibbs AC, Mohan V, Jaeger EP (2005) J Med Chem 48:962–976
Gasteiger J, Sadowski J, Schur J, Selzer P, Steinhauer L, Steinhauer V (1996) J Chem Inf Comput Sci 36:1030–1037
Perkins E, Sun D, Nguyen A, Tulac S, Francesco M, Tavana H, Nguyen H, Tugendreich S, Barthmaier P, Couto J, Yeh E, Thode S, Jarnagin K, Jain AN, Morgans D, Melese T (2001) Cancer Res 61:4175–4183
Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet BK (2002) J Med Chem 45:2213–2221
Wang R, Fang X, Lu Y, Wang S (2004) J Med Chem 47:2977–2980
Jain AN (2006) Curr Protein Pept Sci 7:407–420
Pham TA, Jain AN (2006) J Med Chem 49:5856–5868
Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD (2003) Proteins 52:609–623
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) J Med Chem 47:1739–1749
Wang R, Lai L, Wang S (2002) J Comput Aided Mol Des 16:11–26
Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Arthurs S, Colson AB, Freer ST, Larson V, Luty BA, Marrone T, Rose PW (2000) J Comput Aided Mol Des 14:731–751
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) J Comput Chem 19:1639–1662
Jain AN, Dietterich TG, Lathrop RH, Chapman D, Critchlow RE Jr, Bauer BE, Webster TA, Lozano-Perez T (1994) J Comput Aided Mol Des 8:635–652
Jain AN, Koile K, Chapman D (1994) J Med Chem 37:2315–2327
Jain AN, Harris NL, Park JY (1995) J Med Chem 38:1295–1308
Dietterich TG, Lathrop RH, Lozano-Perez T (1997) Artif Intell 89:31–71
Jain AN (2000) J Comput Aided Mol Des 14:199–213
Fletcher R (1987) Practical methods of optimization, 2nd edn. Wiley-Interscience, New York
Mayo SL, Olafson BD, Goddard WA (1990) J Phys Chem 94:8897–8909
Jain AN (2004) J Med Chem 47:947–961
Lei S, Smith MR (2003) IEEE Trans Softw Eng 29:996–1004
Hawkins PC, Skillman AG, Nicholls A (2007) J Med Chem 50:74–82
Huang N, Shoichet BK, Irwin JJ (2006) J Med Chem 49:6789–6801
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) J Med Chem 49:6177–6196
Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW (2007) J Med Chem 50:726–741
Acknowledgments
The author gratefully acknowledges NIH for partial funding of the work (grant GM070481). Dr. Jain is indebted to Max Cummings for sharing the J&J set, Emanuele Perola for sharing the Vertex Set and performance data, to Bob Clark and Essam Metwally for identifying structural inconsistencies within the original Pham set, and to Ann Cleves for comments on the manuscript. Dr. Jain has a financial interest in BioPharmics LLC, a biotechnology company whose main focus is in the development of methods for computational modeling in drug discovery. Tripos Inc., has exclusive commercial distribution rights for Surflex-Dock, licensed from BioPharmics LLC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jain, A.N. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput Aided Mol Des 21, 281–306 (2007). https://doi.org/10.1007/s10822-007-9114-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-007-9114-2