Abstract
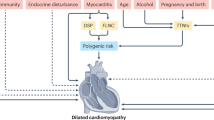
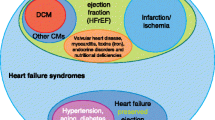
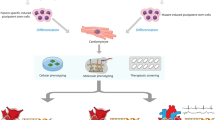
Heart failure is a global health burden responsible for high morbidity and mortality with a prevalence of greater than 60 million individuals worldwide. One of the major causes of heart failure is dilated cardiomyopathy (DCM), characterized by associated systolic dysfunction. During the last few decades, there have been remarkable advances in our understanding about the genetics of dilated cardiomyopathy. The genetic causes were initially thought to be associated with mutations in genes encoding proteins that are localized to cytoskeleton and sarcomere only; however, with the advancement in mechanistic understanding, the roles of ion channels, Z-disc, mitochondria, nuclear proteins, cardiac transcription factors (e.g., NKX-2.5, TBX20, GATA4), and the factors involved in calcium homeostasis have also been identified and found to be implicated in both familial and sporadic DCM cases. During past few years, next-generation sequencing (NGS) has been established as a diagnostic tool for genetic analysis and it has added significantly to the existing candidate gene list for DCM. The animal models have also provided novel insights to develop a better treatment strategy based on phenotype–genotype correlation, epigenetic and phenomic profiling. Most of the DCM biomarkers that are used in routine genetic and clinical testing are structural proteins, but during the last few years, the role of mi-RNA has also emerged as a biomarker due to their accessibility through noninvasive methods. Our increasing genetic knowledge can improve the clinical management of DCM by bringing clinicians and geneticists on one platform, thereby influencing the individualized clinical decision making and leading to precision medicine.



Similar content being viewed by others
References
Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, Dubourg O, Kühl U, Maisch B, McKenna WJ, Monserrat L (2008) Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 29(2):270–276. https://doi.org/10.1093/eurheartj/ehm342
Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB (2006) Contemporary definitions and classification of the cardiomyopathies: an American Heart Association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation 113(14):1807–16. https://doi.org/10.1161/CIRCULATIONAHA.106.174287
Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O’Connell J, Olsen E,Thiene G, Goodwin J, Gyarfas I, Martin I, Nordet P (1996) Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the definition and classification of cardiomyopathies. Circulation 93:841–2. https://doi.org/10.1161/01.cir.93.5.841
Jenni R, Oechslin E, Schneider J, Jost CA, Kaufmann PA (2001) Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: a step towards classification as a distinct cardiomyopathy. Heart. 86(6):666–71. https://doi.org/10.1136/heart.86.6.666
Codd MB, Sugrue DD, Gersh BJ, Melton 3rd LJ (1989) Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy.A population-based study in Olmsted County, Minnesota, 1975–1984. Circulation 80(3):564–72. https://doi.org/10.1161/01.cir.80.3.564
Taylor MR, Carniel E, Mestroni L (2006) Cardiomyopathy, familial dilated. Orphanet J Rare Dis 1:1–8. https://doi.org/10.1186/1750-1172-1-27
Jefferies JL, Towbin JA (2010) Dilated cardiomyopathy. The Lancet 375(9716):752–762. https://doi.org/10.1016/S0140-6736(09)62023-7
Cohn JN, Bristow MR, Chien KR, Colucci WS, Frazier OH, Leinwand LA, Lorell BH, Moss AJ, Sonnenblick EH, Walsh RA, Mockrin SC (1997) Report of the national heart, lung, and blood institute special emphasis panel on heart failure research. Circulation 95(4):766–70. https://doi.org/10.1161/01.cir.95.4.766
Grünig E, Tasman JA, Kücherer H, Franz W, Kübler W, KatusHA (1998) Frequency and phenotypes of familial dilated cardiomyopathy.JAm Col Cardiol. 31(1):186–94. https://doi.org/10.1016/s0735-1097(97)00434-8
Kelly DP, Strauss AW (1994) Inherited cardiomyopathies. NEngl J Med. 330(13):913–9. https://doi.org/10.1056/NEJM199403313301308
Mestroni L, Krajinovic M, Severini GM, Milasin J, Pinamonti B, Rocco C, Vatta M, Falaschi A, Giacca M, Camerini F (1995) Molecular genetics of dilated cardiomyopathies. Eur Heart J. 16(suppl_O):5–9. https://doi.org/10.1093/eurheartj/16.suppl_o.5
Keeling PJ, Gang Y, Smith G, Seo H, Bent SE, Murday V, Caforio AL, McKenna WJ (1995) Familial dilated cardiomyopathy in the United Kingdom. Heart. 73(5):417–21. https://doi.org/10.1136/hrt.73.5.417
Towbin JA, Bowles NE (2002) The failing heart. Nature 415(6868):227–33. https://doi.org/10.1038/415227a
Lipshultz SE, Sleeper LA, Towbin JA, Lowe AM, Orav EJ, Cox GF, Lurie PR, McCoy KL, McDonald MA, Messere JE, Colan SD (2003) The incidence of pediatric cardiomyopathy in two regions of the United States. N Engl J Med. 348(17):1647–55. https://doi.org/10.1056/NEJMoa021715
Towbin JA, Lowe AM, Colan SD, Sleeper LA, Orav EJ, Clunie S, Messere J, Cox GF, Lurie PR, Hsu D, Canter C (2006) Incidence, causes, and outcomes of dilated cardiomyopathy in children. Jama 296(15):1867–76. https://doi.org/10.1001/jama.296.15.1867
Torp A (1978) Incidence of congestive cardiomyopathy. Postgrad Med J. 54(633):435–9. https://doi.org/10.1136/pgmj.54.633.435
Torp A (1981) Incidence of congestive cardiomyopathy. Congestive Cardiomyopathy: Proceedings of a Symposium Held in Kinuna 18–22
Bagger JP, Baandrup U, Rasmussen K, Møller M, Vesterlund T (1984) Cardiomyopathy in western Denmark. Heart 52(3):327–331. https://doi.org/10.1136/hrt.52.3.327
Williams DG, Olsen EG (1985) Prevalence of overt dilated cardiomyopathy in two regions of England. Heart 54(2):153–155. https://doi.org/10.1136/hrt.54.2.153
Rakar S, Sinagra G, Di Lenarda A, Poletti A, Bussani R, Silvestri F, Camerini F (1997) Heart Muscle Disease Study Group. Epidemiology of dilated cardiomyopathy: a prospective post-mortem study of 5252 necropsies. Eur Heart J. 18(1):117–23. https://doi.org/10.1093/oxfordjournals.eurheartj.a015092
Miura K, Nakagawa H, Morikawa Y, Sasayama S, Matsumori A, Hasegawa K, Ohno Y, Tamakoshi A, Kawamura T, Inaba Y (2002) Epidemiology of idiopathic cardiomyopathy in Japan: results from a nationwide survey. Heart 87(2):126–130. https://doi.org/10.1136/heart.87.2.126
Ferencz C, Neill CA (1992) Cardiomyopathy in infancy: observations in an epidemiologic study. Pediatr Cardiol 2:65–71. https://doi.org/10.1007/BF00798206
Arola A, Jokinen E, Ruuskanen O, Saraste M, Pesonen E, Kuusela AL, Tikanoja T, Paavilainen T, Simell O (1997) Epidemiology of idiopathic cardiomyopathies in children and adolescents: a nationwide study in Finland. Am J Epidemiol 146(5):385–393. https://doi.org/10.1093/oxfordjournals.aje.a009291
Nugent AW, Daubeney PE, Chondros P, Carlin JB, Cheung M, Wilkinson LC, Davis AM, Kahler SG, Chow CW, Wilkinson JL, Weintraub RG (2003) The epidemiology of childhood cardiomyopathy in Australia. N. Engl J Med. 348(17):1639–46. https://doi.org/10.1056/NEJMoa021737
Lee TM, Hsu DT, Kantor P, Towbin JA, Ware SM, Colan SD, Chung WK, Jefferies JL, Rossano JW, Castleberry CD, Addonizio LJ (2017) Pediatric cardiomyopathies. Circ Res 121(7):855–73. https://doi.org/10.1161/CIRCRESAHA.116.309386
Petretta M, Pirozzi F, Sasso L, Paglia A, Bonaduce D (2011) Review and metaanalysis of the frequency of familial dilated cardiomyopathy. Am J Cardiol 108(8):1171–1176. https://doi.org/10.1016/j.amjcard.2011.06.022
Ntusi NB, Wonkam A, Shaboodien G, Badri M, Mayosi BM (2011) Frequency and clinical genetics of familial dilated cardiomyopathy in Cape Town: Implications for the evaluation of patients with unexplained cardiomyopathy. S Afr Med J 101(6):394–398
Lakdawala NK, Funke BH, Baxter S, Cirino AL, Roberts AE, Judge DP, Johnson N, Mendelsohn NJ, Morel C, Care M, Chung WK (2012) Genetic testing for dilated cardiomyopathy in clinical practice. J Card Fail. 18(4):296–303. https://doi.org/10.1016/j.cardfail.2012.01.013
van Spaendonck-Zwarts KY, van Rijsingen IA, van den Berg MP, LekanneDeprez RH, Post JG, van Mil AM, Asselbergs FW, Christiaans I, van Langen IM, Wilde AA, de Boer RA. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years' experience. European journal of heart failure. 2013 Jun;15(6):628–36. https://doi.org/10.1093/eurjhf/hft013
Xu L, Zhao L, Yuan F, Jiang WF, Liu H, Li RG, Xu YJ, Zhang M, Fang WY, Qu XK, Yang YQ (2014) GATA6 loss-of-function mutations contribute to familial dilated cardiomyopathy. Int J Mol Med 34(5):1315–1322. https://doi.org/10.3892/ijmm.2014.1896
Zhang XL, Qiu XB, Yuan F, Wang J, Zhao CM, Li RG, Xu L, Xu YJ, Shi HY, Hou XM, Qu XK (2015) TBX5 loss-of-function mutation contributes to familial dilated cardiomyopathy. Biochem Biophys Res Commun. 459(1): 166–71.https://doi.org/10.1016/j.bbrc.2015.02.094
Tayal U, Buchan RJ, Whiffin N, Newsome S, Mazzarotto F, Walsh R, Ware JS, Cook S, Prasad S (2016) 143 Clinical and Genetic Characteristics of Familial Dilated Cardiomyopathy in a Large UK Prospective Cohort. https://doi.org/10.1136/heartjnl-2016-309890.143
Franaszczyk M, Chmielewski P, Truszkowska G, Stawinski P, Michalak E, Rydzanicz M, Sobieszczanska-Malek M, Pollak A, Szczygieł J, Kosinska J, Parulski A (2017) Titin truncating variants in dilated cardiomyopathy–prevalence and genotype-phenotype correlations. PLoS One 12(1):e0169007. https://doi.org/10.1371/journal.pone.0169007
Liu H, Xu YJ, Li RG, Wang ZS, Zhang M, Qu XK, Qiao Q, Li XM, Di RM, Qiu XB, Yang YQ (2019) HAND2 loss-of-function mutation causes familial dilated cardiomyopathy. Eur J Med Genet. 62(9):103540. https://doi.org/10.1016/j.ejmg.2018.09.007
Peña-Peña ML, Ochoa JP, Barriales-Villa R, Cicerchia M, Palomino-Doza J, Salazar-Mendiguchia J, Lamounier A, Trujillo JP, Garcia-Giustiniani D, Fernandez X, Ortiz-Genga M (2020) Clinical utility of genetic testing in patients with dilated cardiomyopathy. Med Clin. https://doi.org/10.1016/j.medcli.2020.05.067
Charron P, Elliott PM, Gimeno JR, Caforio AL, Kaski JP, Tavazzi L, Tendera M, Maupain C, Laroche C, Rubis P, Jurcut R (2018) The Cardiomyopathy Registry of the EURObservational Research Programme of the European Society of Cardiology: baseline data and contemporary management of adult patients with cardiomyopathies. Eur Heart J 39(20):1784–1793. https://doi.org/10.1093/eurheartj/ehx819
Seferović PM, Polovina M, Bauersachs J, Arad M, Gal TB, Lund LH, Felix SB, Arbustini E, Caforio AL, Farmakis D, Filippatos GS (2019) Heart failure in cardiomyopathies: a position paper from the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail 5:553–576. https://doi.org/10.1002/ejhf.1461
Das S, Biswas A, Kapoor M, Seth S, Bhargava B, Rao VR (2015) Epidemiology of cardiomyopathy-A clinical and genetic study of dilated cardiomyopathy: The EPOCH-D study. Journal of the Practice of Cardiovascular Sciences 1(1):30. https://doi.org/10.4103/2395-5414.157562
Kumar D, Vidyapati PML, Kumar M (2017) An Etiological Study of Dilated Cardiomyopathy in Correlation with Clinical, ECG And Echocardiographic Profile. IOSR J Dent Med Sci 16(5):32–36. https://doi.org/10.9790/0853-1605083236
Harikrishnan S, Sanjay G, Anees T, Viswanathan S, Vijayaraghavan G, Bahuleyan CG, Sreedharan M, Biju R, Nair T, Suresh K, Rao AC (2015) Clinical presentation, management, in-hospital and 90-day outcomes of heart failure patients in Trivandrum, Kerala, India: the Trivandrum Heart Failure Registry. Eur J Heart Fail 17(8):794–800. https://doi.org/10.1002/ejhf.283
Towbin JA, Hejtmancik JF, Brink P, Gelb B, Zhu XM, Chamberlain JS, McCabe ER, Swift M (1993) X-linked dilated cardiomyopathy. Molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation 87(6):1854–65. https://doi.org/10.1161/01.cir.87.6.1854
Zhang D, Mott JL, Farrar P, Ryerse JS, Chang SW, Stevens M, Denniger G, Zassenhaus HP (2003) Mitochondrial DNA mutations activate the mitochondrial apoptotic pathway and cause dilated cardiomyopathy. Cardiovasc Res. 57(1):147–57. https://doi.org/10.1016/s0008-6363(02)00695-8
Lloyd DF, Vara R, Mathur S (2017) Cardiac manifestations of inherited metabolic disease in children. Pediatrics International 59(5):525–9. https://doi.org/10.1111/ped.13272
Chubb H, Simpson JM (2012) The use of Z-scores in paediatric cardiology.Ann Pediatr Cardiol. 5(2):179.https://doi.org/10.4103/0974-2069.99622
Mestroni L, Rocco C, Gregori D, Sinagra G, Di Lenarda A, Miocic S, Vatta M, Pinamonti B, Muntoni F, Caforio AL, McKenna WJ (1999) Familial dilated cardiomyopathy: evidence for genetic and phenotypic heterogeneity.JAm Coll Cardiol. 34(1):181–190. https://doi.org/10.1016/s0735-1097(99)00172-2
Elliott P (2000) Diagnosis and management of dilated cardiomyopathy. Heart 84(1):106. https://doi.org/10.1136/heart.84.1.106
Sinagra G, Merlo M, Pinamonti B (2019) Dilated Cardiomyopathy. Springer. https://doi.org/10.1007/978-3-030-13864-6
Dec GW, Fuster V (1994) Idiopathic dilated cardiomyopathy. N Engl J Med. 331(23):1564–75.https://doi.org/10.1056/NEJM199412083312307
Arbustini E, Di Toro A, Giuliani L, Favalli V, Narula N, Grasso M (2018) Cardiac phenotypes in hereditary muscle disorders: JACC state-of-the-art review.JAm Coll Cardiol.72(20):2485–506. https://doi.org/10.1016/j.jacc.2018.08.2182
Goldenberg J, Ferraz MB, Pessoa AP, Fonseca AS, Carvalho AC, Hilario MO, Atra E (1992) Symptomatic cardiac involvement in juvenile rheumatoid arthritis. Int J Cardiol. 34(1):57–62. https://doi.org/10.1016/0167-5273(92)90082-e
Hantson P (2019) Mechanisms of toxic cardiomyopathy. Clin Toxicol. 57(1):1–9. https://doi.org/10.1080/15563650.2018.1497172
Vikhorev PG, Smoktunowicz N, Munster AB, Kostin S, Montgiraud C, Messer AE, Toliat MR, Li A, Dos Remedios CG, Lal S, Blair CA (2017) Abnormal contractility in human heart myofibrils from patients with dilated cardiomyopathy due to mutations in TTN and contractile protein genes. Sci Rep 1;7(1):1–1. https://doi.org/10.1038/s41598-017-13675-8
Vang S, Corydon TJ, Børglum AD, Scott MD, Frydman J, Mogensen J, Gregersen N, Bross P (2005) Actin mutations in hypertrophic and dilated cardiomyopathy cause inefficient protein folding and perturbed filament formation. The FEBS Journal 272(8):2037–49. https://doi.org/10.1111/j.1742-4658.2005.04630.x
Gautel M, Zuffardi O, Freiburg A, Labeit S (1995) Phosphorylation switches specific for the cardiac isoform of myosin binding protein‐C: a modulator of cardiac contraction? The EMBO journal 14(9):1952–60. https://doi.org/10.1002/j.1460-2075.1995.tb07187.x
Daehmlow S, Erdmann J, Knueppel T, Gille C, Froemmel C, Hummel M, Hetzer R, Regitz-Zagrosek V (2002) Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem Biophys Res Commun 298(1):116–120. https://doi.org/10.1016/S0006-291X(02)02374-4
Li X, Luo R, Gu H, Deng Y, Xu X, Wu X, Hua W (2014) Cardiac troponin T (TNNT2) mutations in Chinese dilated cardiomyopathy patients. Biomed Research Int.2014. https://doi.org/10.1155/2014/907360
Mogensen J, Murphy RT, Shaw T, Bahl A, Redwood C, Watkins H, Burke M, Elliott PM, WJ, (2004) Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol 44(10):2033–2040. https://doi.org/10.1016/j.jacc.2004.08.027
Marques M, Pinto J, Moraes A, Sorenson M, Silva J, Oliveira G (2015) Structural Behavior of Cardiac Troponin C Variants Present in Cardiomyopathic Patients. Biophys J. 108(2):213a. https://doi.org/10.1002/j.1460-2075.1995.tb07187.x
Pan S, Sommese RF, Sallam KI, Nag S, Sutton S, Miller SM, Spudich JA, Ruppel KM, Ashley EA (2015) Establishing disease causality for a novel gene variant in familial dilated cardiomyopathy using a functional in-vitro assay of regulated thin filaments and human cardiac myosin. BMC Med Genet 16(1):1–7. https://doi.org/10.1186/s12881-015-0243-5
Li D, Czernuszewicz GZ, Gonzalez O, Tapscott T, Karibe A, Durand JB, Brugada R, Hill R, Gregoritch JM, Anderson JL, Quiñones M (2001) Novel cardiac troponin T mutation as a cause of familial dilated cardiomyopathy. Circulation 104(18):2188–2193. https://doi.org/10.1161/hc4301.098285
Lakdawala NK, Dellefave L, Redwood CS, Sparks E, Cirino AL, Depalma S, Colan SD, Funke B, Zimmerman RS, Robinson P, Watkins H (2010) Familial dilated cardiomyopathy caused by an alpha-tropomyosin mutation: the distinctive natural history of sarcomeric dilated cardiomyopathy. J Am Coll Cardiol 55(4):320–9. https://doi.org/10.1016/j.jacc.2009.11.017
van de Meerakker JB, Christiaans I, Barnett P, Deprez RH, Ilgun A, Mook OR, Mannens MM, Lam J, Wilde AA, Moorman AF, Postma AV (2013) A novel alpha-tropomyosin mutation associates with dilated and non-compaction cardiomyopathy and diminishes actin binding. BiochimicaetBiophysicaActa (BBA)-Molecular Cell Research 1833(4):833–9. https://doi.org/10.1016/j.bbamcr.2012.11.003
Olson TM, Kishimoto NY, Whitby FG, Michels VV (2001) Mutations that alter the surface charge of alpha-tropomyosinare associated with dilated cardiomyopathy. J Moll Cell Cardiol. 33(4):723–32. https://doi.org/10.1006/jmcc.2000.1339
Mirza M, Robinson P, Kremneva E, Nikolaeva O, Watkins H, Levitsky D, Redwood C, Mohammed EM, Marston S (2007) The effect of mutations in α-tropomyosin (E40K and E54K) that cause familial dilated cardiomyopathy on the regulatory mechanism of cardiac muscle thin filaments. J Biol Chem. 282(18):13487–97. https://doi.org/10.1074/jbc.M701071200
Whitby FG, Phillips Jr GN (2000) Crystal structure of tropomyosin at 7 Ångstroms resolution. Proteins: Structure, Function, and Bioinformatics 38(1):49–59. https://onlinelibrary.wiley.com/doi/full/10.1002/%28SICI%291097-0134%2820000101%2938%3A1%3C49%3A%3AAID-PROT6%3E3.0.CO%3B2-B
Henderson CA, Gomez CG, Novak SM, Mi‐Mi L, Gregorio CC (2011) Overview of the muscle cytoskeleton. Compr Physiol 7(3):891–944. https://doi.org/10.1002/cphy.c160033
Tsikitis M, Galata Z, Mavroidis M, Psarras S, Capetanaki Y (2018) Intermediate filaments in cardiomyopathy. Biophys Rev. 10(4):1007–31. https://doi.org/10.1007/s12551-018-0443-2
Ehler E (2018) Actin-associated proteins and cardiomyopathy—the ‘unknown’beyond troponin and tropomyosin. Biophys Rev. 10(4):1121–8. https://doi.org/10.1007/s12551-018-0428-1
Granzier HL, Labeit S (2004) The giant protein titin: a major player in myocardial mechanics, signaling, and disease Circ Res 94:284–295. https://doi.org/10.1161/01.RES.0000117769.88862.F8
Tabish AM, Azzimato V, Alexiadis A, Buyandelger B, Knöll R (2017) Genetic epidemiology of titin-truncating variants in the etiology of dilated cardiomyopathy.Biophys Rev. 9(3):207–23. https://doi.org/10.1007/s12551-017-0265-7
Frank D, Frey N (2011) Cardiac Z-disc signaling network. J Biol Chem 286(12):9897–904.https://doi.org/10.1074/jbc.R110.174268
Despond EA, Dawson JF (2018) Classifying cardiac actin mutations associated with hypertrophic cardiomyopathy. Front Physiol. 9:405. https://doi.org/10.3389/fphys.2018.00405
Frank D, Rangrez AY, Friedrich C, Dittmann S, Stallmeyer B, Yadav P, Bernt A, Schulze-Bahr E, Borlepawar A, Zimmermann WH, Peischard S (2019) Cardiac α-actin (ACTC1) gene mutation causes atrial-septal defects associated with late-onset dilated cardiomyopathy. Circulation: Genomic and Precision Medicine 12(8):e002491. https://doi.org/10.1161/CIRCGEN.119.002491
Rangrez AY, Kilian L, Stiebeling K, Dittmann S, Schulze-Bahr E, Frey N, Frank D (2019) A cardiac α-actin (ACTC1) p. Gly247Asp mutation inhibits SRF-signaling in vitro in neonatal rat cardiomyocytes. Biochem Biophys Res Commun. 518(3):500–5. https://doi.org/10.1016/j.bbrc.2019.08.081
Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL (2012) Truncations of titin causing dilated cardiomyopathy. N Engl J Med 366(7):619–628. https://doi.org/10.1056/NEJMoa1110186
Pugh TJ, Kelly MA, Gowrisankar S, Hynes E, Seidman MA, Baxter SM, Bowser M, Harrison B, Aaron D, Mahanta LM, Lakdawala NK (2014) The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 16(8):601–8. https://doi.org/10.1038/gim.2013.204
Fatkin D, Huttner IG (2017) Titin-truncating mutations in dilated cardiomyopathy: the long and short of it. Curr Opin Cardiol. 32(3):232–8. https://doi.org/10.1097/HCO.0000000000000382
Vikhorev PG, Vikhoreva NN, Yeung W, Li A, Lal S, Dos Remedios CG, Blair CA, Guglin M, Campbell KS, Yacoub MH, de Tombe P (2020) Titin-truncating mutations associated with dilated cardiomyopathy alter length-dependent activation and its modulation via phosphorylation. Cardiovasc Res. https://doi.org/10.1093/cvr/cvaa316
Purevjav E, Arimura T, Augustin S, Huby AC, Takagi K, Nunoda S, Kearney DL, Taylor MD, Terasaki F, Bos JM, Ommen SR (2012) Molecular basis for clinical heterogeneity in inherited cardiomyopathies due to myopalladin mutations. Hum Mol Genet. 21(9):2039–53. https://doi.org/10.1093/hmg/dds022
Perrot A, Tomasov P, Villard E, Faludi R, Melacini P, Lossie J, Lohmann N, Richard P, De Bortoli M, Angelini A, Varga-Szemes A (2016) Mutations in NEBL encoding the cardiac Z-disk protein nebulette are associated with various cardiomyopathies. Arch Med Sci: AMS 12(2):263. https://doi.org/10.5114/aoms.2016.59250
Begay RL, Graw SL, Sinagra G, Asimaki A, Rowland TJ, Slavov DB, Gowan K, Jones KL, Brun F, Merlo M, Miani D (2018) Filamin C truncation mutations are associated with arrhythmogenic dilated cardiomyopathy and changes in the cell–cell adhesion structures. JACC: Clinic Electrophysiol. 4(4):504–14. https://doi.org/10.1016/j.jacep.2017.12.003
Filomena MC, Yamamoto DL, Carullo P, Piroddi N, Tesi C, Scellini B, Crispino R, Zhang J, Knöll R, Polishchuk R, Poggesi C (2018) Myopalladin is upregulated in dilated cardiomyopathies patients and myopalladin knockout mice develop cardiac dilation and dysfunction following pressure overload. J Mol Cell Cardiol 120:43. https://doi.org/10.1016/j.yjmcc.2018.05.129
Good JM, Fellmann F, Bhuiyan ZA, Rotman S, Pruvot E, Schläpfer J (2020) ACTN2 variant associated with a cardiac phenotype suggestive of left-dominant arrhythmogenic cardiomyopathy. HeartRhythm case reports 6(1):15. https://doi.org/10.1016/j.hrcr.2019.10.001
Mohapatra B, Jimenez S, Lin JH, Bowles KR, Coveler KJ, Marx JG, Chrisco MA, Murphy RT, Lurie PR, Schwartz RJ, Elliott PM (2013) Mutations in the muscle LIM protein and α-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 80(1–2):207–15.https://doi.org/10.1016/s1096-7192(03)00142-2
Hayashi T, Arimura T, Itoh-Satoh M, Ueda K, Hohda S, Inagaki N, Takahashi M, Hori H, Yasunami M, Nishi H, Koga Y (2004) Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol 44(11):2192–2201. https://doi.org/10.1016/j.jacc.2004.08.058
Duboscq-Bidot L, Xu P, Charron P, Neyroud N, Dilanian G, MillaireA, Bors V, Komajda M, Villard E (2008) Mutations in the Z-band protein myopalladin gene and idiopathic dilated cardiomyopathy. Cardiovas Res. 77(1):118–25. https://doi.org/10.1093/cvr/cvm015
Ehmsen J, Poon E, Davies K (2002) The dystrophin-associated protein complex. J Cell Sci 115(14):2801–2803
Constantin B (2014) Dystrophin complex functions as a scaffold for signalling proteins. Biochimica et Biophysica Acta (BBA)-Biomembranes 1838(2):635–42. https://doi.org/10.1074/jbc.R110.174268
Abdallah AM, Carlus SJ, Al-Mazroea AH, Alluqmani M, Almohammadi Y, Bhuiyan ZA, Al-Harbi KM (2019) Digenic inheritance of LAMA4 and MYH7 mutations in patient with infantile dilated cardiomyopathy. Medicina 55(1):17. https://doi.org/10.3390/medicina55010017
Knoll R, Postel R et al (2007) Laminin-alpha 4 and Integrin-Linked Kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Identification and Characterization of Novel Genes by Reverse and Forward Genetics in Zebrafish 116:47. https://doi.org/10.1161/circulationaha.107.689984
Murakami T, Hayashi YK, Noguchi S, Ogawa M, Nonaka I, Tanabe Y, Ogino M, Takada F, Eriguchi M, Kotooka N, Campbell KP (2006) Fukutin gene mutations cause dilated cardiomyopathy with minimal muscle weakness. Annals of Neurology: Official Journal of the American Neurological Association and the Child Neurology Society 60(5):597–602. https://doi.org/10.1002/ana.20973
Ohno S (2016) The genetic background of arrhythmogenic right ventricular cardiomyopathy. J Arrhythm.32(5):398–403. https://doi.org/10.1016/j.joa.2016.01.006
Brun F, Barnes CV, Sinagra G, Slavov D, Barbati G, Zhu X et al (2014) Familial Cardiomyopathy Registry. Titin and desmosomal genes in the natural history of arrhythmogenic right ventricular cardiomyopathy. J Med Genet 51:669–676. https://doi.org/10.1136/jmedgenet-2014-102591
Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura KI, Utsumi H, Hamasaki N, Takeshita A (2011) Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res. 88(5):529–35.https://doi.org/10.1161/01.res.88.5.529
Philipp U, Broschk C, Vollmar A, Distl O (2007) Evaluation of tafazzin as candidate for dilated cardiomyopathy in Irish wolfhounds. J Hered 98(5):506–509. https://doi.org/10.1093/jhered/esm045
Zhu M (2020) Deletion of tafazzin in cardiomyocytes results in dilated cardiomyopathy (Doctoral dissertation, UC San Diego)
Szent-Györgyi AG (1975) Calcium regulation of muscle contraction. Biophys J. 15(7):707–23. https://doi.org/10.1016/S0006-3495(75)85849-8
Haghighi K, Kolokathis F, Pater L, Lynch RA, Asahi M, Gramolini AO, Fan GC, Tsiapras D, Hahn HS, Adamopoulos S, Liggett SB (2003) Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Investig 111(6):869–876. https://doi.org/10.1172/JCI17892
Fish M, Shaboodien G, Kraus S, Sliwa K, Seidman CE, Burke MA, Crotti L, Schwartz PJ, Mayosi BM (2016) Corrigendum: Mutation analysis of the phospholamban gene in 315 South Africans with dilated, hypertrophic, peripartum and arrhythmogenic right ventricular cardiomyopathies Sci Rep 6. https://doi.org/10.1038/srep22235
Sipido KR, Vangheluwe P (2010) Targeting sarcoplasmic reticulum Ca2+ uptake to improve heart failure: hit or miss. https://doi.org/10.1161/CIRCRESAHA.109.210740
Li W, Yin L, Shen C, Hu K, Ge J, Sun A (2018) SCN5A variants: association with cardiac disorders. Front Physiol. 9:1372.https://doi.org/10.3389/fphys.2018.01372
Bienengraeber M, Olson TM, Selivanov VA, Kathmann EC, O'Cochlain F, Gao F, Karger AB, Ballew JD, Hodgson DM, Zingman LV, Pang YP (2004) ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic K ATP channel gating. Nat Genet. 36(4):382–7.https://doi.org/10.1038/ng1329
El-Battrawy I, Zhao Z, Lan H, Li X, Yücel G, Lang S, Sattler K, Schünemann JD, Zimmermann WH, Cyganek L, UtikalJ (2018) Ion channel dysfunctions in dilated cardiomyopathy in limb-girdle muscular dystrophy. Circ: Genom Precis Med. 11(3):e001893. https://doi.org/10.1161/CIRCGEN.117.001893
Stroud MJ (2018) Linker of nucleoskeleton and cytoskeleton complex proteins in cardiomyopathy. Biophys Rev. 10(4):1033–51.https://doi.org/10.1007/s12551-018-0431-6
Hershberger RE, Morales A (1993) LMNA-related dilated cardiomyopathy. In: GeneReviews®. University of Washington, Seattle, Seattle (WA)
Hasselberg NE, Haland TF, Saberniak J, Brekke PH, Berge KE, Leren TP, Edvardsen T, Haugaa KH (2018) Lamin A/C cardiomyopathy: young onset, high penetrance, and frequent need for heart transplantation. Eur Heart J. 39(10):853–60. https://doi.org/10.1093/eurheartj/ehx596
Sinagra G, Dal Ferro M, Merlo M (2017) Lamin A/C cardiomyopathy: cutting edge to personalized medicine 10(6):e002004.https://doi.org/10.1161/CIRCGENETICS.117.002004
Berry DA, Keogh A, Remedios CG (2001) Nuclear membrane proteins in failing human dilated cardiomyopathy. PROTEOMICS: International Edition 1(12):1507–12: https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/abs/10.1002/1615-9861%28200111%291%3A12%3C1507%3A%3AAID-PROT1507%3E3.0.CO%3B2-Z
Arbustini E, Pilotto A, Repetto A, Grasso M, Negri A, Diegoli M, Campana C, Scelsi L, Baldini E, Gavazzi A, Tavazzi L (2002) Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J Am Coll Cardiol. 39(6):981–90. https://doi.org/10.1016/s0735-1097(02)01724-2
McNally EM, Golbus JR, Puckelwartz MJ (2013) Genetic mutations and mechanisms in dilated cardiomyopathy. J Clinic Investig. 123(1):19–26 https://doi.org/10.1172/JCI62862
Li J, Liu WD, Yang ZL, Yuan F, Xu L, Li RG, Yang YQ (2014) Prevalence and spectrum of GATA4 mutations associated with sporadic dilated cardiomyopathy. Gene 548(2):174–81.https://doi.org/10.1016/j.gene.2014.07.022
Li RG, Li L, Qiu XB, Yuan F, Xu L, Li X, Xu YJ, Jiang WF, Jiang JQ, Liu X, Fang WY (2013) GATA4 loss-of-function mutation underlies familial dilated cardiomyopathy. Biochem Biophys Res Commun. 439(4):591–6. https://doi.org/10.1016/j.bbrc.2013.09.023
Zhao L, Xu JH, Xu WJ, Yu H, Wang Q, Zheng HZ, Jiang WF, Jiang JF, Yang YQ (2014) A novel GATA4 loss-of-function mutation responsible for familial dilated cardiomyopathy. Int J Mol Med. 33(3):654–60. https://doi.org/10.3892/ijmm.2013.1600
Mittal A, Sharma R, Prasad R, Bahl A, Khullar M (2016) Role of cardiac TBX20 in dilated cardiomyopathy. Mol Cell Biochem. 414(1):129–36. https://doi.org/10.1007/s11010-016-2666-5
Zhou YM, Dai XY, Huang RT, Xue S, Xu YJ, Qiu XB, Yang YQ (2016) A novel TBX20 loss-of-function mutation contributes to adult-onset dilated cardiomyopathy or congenital atrial septal defect. Mol Med Rep. 14(4):3307–14. https://doi.org/10.3892/mmr.2016.5609
Yuan F, Qiu ZH, Wang XH, Sun YM, Wang J, Li RG, Liu H, Zhang M, Shi HY, Zhao L, Jiang WF (2018) MEF2C loss-of-function mutation associated with familial dilated cardiomyopathy. Clinic Chem Lab Med. (CCLM) 56(3):502–11. https://doi.org/10.1515/cclm-2017-0461
Huang J, Lu MM, Cheng L, Yuan LJ, Zhu X, Stout AL, Chen M, Li J, Parmacek MS (2009) Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc Natl Acad Sci. 106(44):18734–9. https://doi.org/10.1073/pnas.0910749106
Torrado M, López E, Centeno A, Medrano C, Castro-Beiras A, Mikhailov AT (2003) Myocardin mRNA is augmented in the failing myocardium: expression profiling in the porcine model and human dilated cardiomyopathy. J Mol Med. 81(9):566–77. https://doi.org/10.1007/s00109-003-0470-7
Auguste G, Gurha P, Lombardi R, Coarfa C, Willerson JT, Marian AJ (2018) Suppression of activated FOXO transcription factors in the heart prolongs survival in a mouse model of laminopathies. Circ Res. 122(5):678–92. https://doi.org/10.1161/CIRCRESAHA.117.312052
Xu JH, Gu JY, Guo YH, Zhang H, Qiu XB, Li RG, Shi HY, Liu H, Yang XX, Xu YJ, Qu XK (2017) Prevalence and spectrum of NKX2–5 mutations associated with sporadic adult-onset dilated cardiomyopathy. Int Heart J. 58(4):521–9. https://doi.org/10.1536/ihj.16-440
Yuan F, Qiu XB, Li RG, Qu XK, Wang J, Xu YJ, Liu X, Fang WY, Yang YQ, Liao DN (2015) A novel NKX2–5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int J Mol Med. 35(2):478–86. https://doi.org/10.3892/ijmm.2014.2029
Sveinbjornsson G, Olafsdottir EF, Thorolfsdottir RB, Davidsson OB, Helgadottir A, Jonasdottir A, Jonasdottir A, Bjornsson E, Jensson BO, Arnadottir GA, Kristinsdottir H (2018) Variants in NKX2–5 and FLNC cause dilated cardiomyopathy and sudden cardiac death. Circ: Genom Precis Med. 11(8):e002151. https://doi.org/10.1161/CIRCGEN.117.002151
Parlakian A, Charvet C, Escoubet B, Mericskay M, Molkentin JD, Gary-Bobo G, De Windt LJ, Ludosky MA, Paulin D, Daegelen D, Tuil D (2005) Clinical perspective. Circulation 112(19):2930–9. https://doi.org/10.1161/CIRCULATIONAHA.105.533778
Miao L, Li J, Li J, Tian X, Lu Y, Hu S, Shieh D, Kanai R, Zhou BY, Zhou B, Liu J (2018) Notch signaling regulates Hey2 expression in a spatiotemporal dependent manner during cardiac morphogenesis and trabecular specification. Sci Rep. 8(1):1–4. https://doi.org/10.1038/s41598-018-20917-w
Li RG, Li L, Qiu XB, Yuan F, Xu L, Li X, Xu YJ, Jiang WF, Jiang JQ, Liu X, Fang WY (2013) GATA4 loss-of-function mutation underlies familial dilated cardiomyopathy. Biochem Biophys Res Commun 439(4):591–596. https://doi.org/10.1016/j.bbrc.2013.09.023
Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I, Sperling SR (2011) The cardiac transcription network modulated by Gata4, Mef2a, Nkx2. 5, Srf, histone modifications, and microRNAs. PLoS Genet. 7(2):e1001313. https://doi.org/10.1371/journal.pgen.1001313
Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, Mackay JP (2007) Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 81(2):280–91. https://doi.org/10.1086/519530
Xu JH, Gu JY, Guo YH, Zhang H, Qiu XB, Li RG, Shi HY, Liu H, Yang XX, Xu YJ, Qu XK (2017) Prevalence and spectrum of NKX2–5 mutations associated with sporadic adult-onset dilated cardiomyopathy. Int Heart J. 58(4):521–9. https://doi.org/10.1536/ihj.16-440
Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, Witt CC, Becker K, Labeit S, Granzier HL (2004) Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation 110(2):155–62. https://doi.org/10.1161/01.CIR.0000135591.37759.AF
Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V (2012) RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 18(5):766–73. https://doi.org/10.1038/nm.2693
Li D, Morales A, Gonzalez-Quintana J, Norton N, Siegfried JD, Hofmeyer M, Hershberger RE (2010) Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci 3(3):90–97. https://doi.org/10.1111/j.1752-8062.2010.00198.x
Refaat MM, Lubitz SA, Makino S, Islam Z, Frangiskakis JM, Mehdi H, Gutmann R, Zhang ML, Bloom HL, MacRae CA, Dudley SC (2012) Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm 9(3):390–396. https://doi.org/10.1016/j.hrthm.2011.10.016
Zhu C, Yin Z, Tan B, Guo W (2017) Insulin regulates titin pre-mRNA splicing through the PI3K-Akt-mTOR kinase axis in a RBM20-dependent manner. BiochimicaetBiophysicaActa (BBA)-Molecular Basis of Disease 1863(9):2363–71. https://doi.org/10.1016/j.bbadis.2017.06.023
Tharp CA, Haywood ME, Sbaizero O, Taylor MR, Mestroni L (2019) The giant protein titin’s role in cardiomyopathy: genetic, transcriptional, and post-translational modifications of TTN and their contribution to cardiac disease. Front Physiol. 10:1436. https://doi.org/10.3389/fphys.2019.01436
Lefeber DJ, de Brouwer AP, Morava E, Riemersma M, Schuurs-Hoeijmakers JH, Absmanner B, Verrijp K, van den Akker WM, Huijben K, Steenbergen G, van Reeuwijk J (2011) Autosomal recessive dilated cardiomyopathy due to DOLK mutations results from abnormal dystroglycan O-mannosylation. PLoS Genet 7(12):e1002427. https://doi.org/10.1371/journal.pgen.1002427
Yu J, Zeng C, Wang Y (2019) Epigenetics in dilated cardiomyopathy. Curr Opin Cardiol 34(3):260. https://doi.org/10.1097/hco.0000000000000616
Gi WT, Haas J, Sedaghat-Hamedani F, Kayvanpour E, Tappu R, Lehmann DH, ShirvaniSamani O, Wisdom M, Keller A, Katus HA, Meder B (2020) Epigenetic Regulation of Alternative mRNA Splicing in Dilated Cardiomyopathy. J Clinic Med 9(5):1499. https://doi.org/10.3390/jcm9051499
Haas J, Frese KS, Park YJ, Keller A, Vogel B, Lindroth AM, Weichenhan D, Franke J, Fischer S, Bauer A, Marquart S (2013) Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol Med 5(3):413–429. https://doi.org/10.1002/emmm.201201553
Gramlich M, Pane LS, Zhou Q, Chen Z, Murgia M, Schötterl S, Goedel A, Metzger K, Brade T, Parrotta E, Schaller M (2015) Antisense‐mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol Med. 7(5):562–76. https://doi.org/10.15252/emmm.201505047
Ito E, Miyagawa S, Fukushima S, Yoshikawa Y, Saito S, Saito T, Harada A, Takeda M, Kashiyama N, Nakamura Y, Shiozaki M (2017) Histone modification is correlated with reverse left ventricular remodeling in nonischemic dilated cardiomyopathy. Annals Thorac Surg. 104(5):1531–9. https://doi.org/10.1016/j.athoracsur.2017.04.046
Jiang DS, Yi X, Li R, Su YS, Wang J, Chen ML, Liu LG, Hu M, Cheng C, Zheng P, Zhu XH (2017) The histone methyltransferase mixed lineage leukemia (MLL) 3 may play a potential role in clinical dilated cardiomyopathy. Mol Med. 23(1):196–203. https://doi.org/10.2119/molmed.2017.00012
Theis JL, Sharpe KM, Matsumoto ME, Chai HS, Nair AA, Theis JD, De Andrade M, Wieben ED, Michels VV, Olson TM (2011) Homozygosity mapping and exome sequencing reveal GATAD1 mutation in autosomal recessive dilated cardiomyopathy. Circulation: Cardiovascular Genetics 4(6):585–94. https://doi.org/10.1161/CIRCGENETICS.111.961052
Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN (2007) Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes & Development 21(14):1790–802. https://doi.org/10.1007/s12551-018-0428-1
Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN (2008) microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 22(23):3242–3254. https://doi.org/10.1101/gad.1738708
Van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN (2006) A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci. 103(48):18255–60. https://doi.org/10.1073/pnas.0608791103
Hamada H, Suzuki M, Yuasa S, Mimura N, Shinozuka N, Takada Y, Suzuki M, Nishino T, Nakaya H, Koseki H, Aoe T (2004) Dilated cardiomyopathy caused by aberrant endoplasmic reticulum quality control in mutant KDEL receptor transgenic mice. Mol Cel Biol. 24(18):8007–17. https://doi.org/10.1128/MCB.24.18.8007-8017.2004
Li D, Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Partain J, Nixon RR, Allen CN, Irwin RP, Jakobs PM (2006) Mutations of presenilin genes in dilated cardiomyopathy and heart failure. Am J Hum Genet 79(6):1030–1039. https://doi.org/10.1086/509900
Gianni D, Li A, Tesco G, McKay KM, Moore J, Raygor K, Rota M, Gwathmey JK, Dec GW, Aretz T, Leri A (2010) Protein aggregates and novel presenilin gene variants in idiopathic dilated cardiomyopathy. Circulation 121(10):1216. https://doi.org/10.1161/CIRCULATIONAHA.109.879510
Dhandapany PS, Razzaque MA, Muthusami U, Kunnoth S, Edwards JJ, Mulero-Navarro S, Riess I, Pardo S, Sheng J, Rani DS, Rani B (2014) RAF1 mutations in childhood-onset dilated cardiomyopathy. Nat Genet 46(6):635–639. https://doi.org/10.1038/ng.2963
Zhao Y, Feng Y, Zhang YM, Ding XX, Song YZ, Zhang AM, Liu L, Zhang H, Ding JH, Xia XS (2015) Targeted next-generation sequencing of candidate genes reveals novel mutations in patients with dilated cardiomyopathy. Int J Mol Med 36(6):1479–86. https://doi.org/10.3892/ijmm.2015.2361
Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, Feng Z, Müller S, KayvanpourE, Vogel B, Sedaghat-Hamedani F (2015) Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 36(18):1123–35. https://doi.org/10.1093/eurheartj/ehu301
Norton N, Li D, Rieder MJ, Siegfried JD, Rampersaud E, Züchner S, Mangos S, Gonzalez-Quintana J, Wang L, McGee S, Reiser J (2011) Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 88(3):273–82. https://doi.org/10.1016/j.ajhg.2011.01.016
Arimura T, Ishikawa T, Nunoda S, Kawai S, Kimura A (2011) Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum Mutat. 32(12):1481–91. https://doi.org/10.1002/humu.21603
Campbell N, Sinagra G, Jones KL, Slavov D, Gowan K, Merlo M, Carniel E, Fain PR, Aragona P, Di Lenarda A, Mestroni L (2013) Whole exome sequencing identifies a troponin T mutation hot spot in familial dilated cardiomyopathy. PloS One 8(10):e78104. https://doi.org/10.1371/journal.pone.0078104
Zhang M, Chen J, Si D, Zheng Y, Jiao H, Feng Z, Hu Z, Duan R (2014) Whole exome sequencing identifies a novel EMD mutation in a Chinese family with dilated cardiomyopathy. BMC medical genetics.15(1):1–7. https://doi.org/10.1186/1471-2350-15-77
Akinrinade O, Ollila L, Vattulainen S, Tallila J, Gentile M, Salmenperä P, Koillinen H, Kaartinen M, Nieminen MS, Myllykangas S, Alastalo TP zhao (2015) Genetics and genotype–phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur Heart J. 36(34):2327–37. https://doi.org/10.1093/eurheartj/ehv253
Truszkowska GT, Bilińska ZT, Muchowicz A, Pollak A, Biernacka A, Kozar-Kamińska K, Stawiński P, Gasperowicz P, Kosińska J, Zieliński T, Płoski R (2017) Homozygous truncating mutation in NRAP gene identified by whole exome sequencing in a patient with dilated cardiomyopathy. Sci Rep. 7(1):1–5. https://doi.org/10.1038/s41598-017-03189-8
Dvornikov AV, de Tombe PP, Xu X (2018) Phenotyping cardiomyopathy in adult zebrafish. Prog Biophys Mol Biol 138:116–125. https://doi.org/10.1016/j.pbiomolbio.2018.05.013
JD Federspiel P Tandon CM Wilczewski L Wasson LE Herring SS Venkatesh IM Cristea FL Conlon 2019 Conservation and divergence of protein pathways in the vertebrate heart PLoS Biol 17 9 e3000437 https://doi.org/10.1371/journal.pbio.3000437
Ding Y, Bu H, Xu X (2020) Modeling Inherited Cardiomyopathies in Adult Zebrafish for Precision Medicine. Front Physiol 11:1526. https://doi.org/10.3389/fphys.2020.599244
Arber S, Hunter JJ, Ross Jr J, Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P (1997) MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 88(3):393–403. https://doi.org/10.1016/s0092-8674(00)81878-4
Minamisawa S, Hoshijima M, Chu G, Ward CA, Frank K, Gu Y, Martone ME, Wang Y, Ross Jr J, Kranias EG, Giles WR (1999) Chronic phospholamban–sarcoplasmic reticulum calcium ATPase interaction is the critical calcium cycling defect in dilated cardiomyopathy. Cell 99(3):313–22. https://doi.org/10.1016/s0092-8674(00)81662-1
Wang X, Osinska H, Dorn GW, Nieman M, Lorenz JN, Gerdes AM, Witt S, Kimball T, Gulick J, Robbins J (2001) Mouse model of desmin-related cardiomyopathy. Circulation 103(19):2402–7. https://doi.org/10.1161/01.cir.103.19.2402
Milner DJ, Taffet GE, Wang X, Pham T, Tamura T, Hartley C, Gerdes MA, Capetanaki Y (1999) The absence of desmin leads to cardiomyocyte hypertrophy and cardiac dilation with compromised systolic function. J Moll Cell Cardiol. 31(11):2063–76. https://doi.org/10.1006/jmcc.1999.1037
McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H (1999) Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Investig. 104(9):1235–44. https://doi.org/10.1172/JCI7377
Sussman MA, Welch S, Cambon N, Klevitsky R, Hewett TE, Price R, Witt SA, Kimball TR (1998) Myofibril degeneration caused by tropomodulin overexpression leads to dilated cardiomyopathy in juvenile mice. J Clin Investig 101(1):51–61. https://doi.org/10.1172/JCI1167
Megeney LA, Kablar B, Perry RL, Ying C, May L, Rudnicki MA (1999) Severe cardiomyopathy in mice lacking dystrophin and MyoD. Proc Natl Acad Sci 96(1):220–225. https://doi.org/10.1073/pnas.96.1.220
Hack AA, Ly CT, Jiang F, Clendenin CJ, Sigrist KS, Wollmann RL, McNally EM (1998) γ-Sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J Cell Biol 142(5):1279–1287. https://doi.org/10.1083/jcb.142.5.1279
Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, Straub V, Barresi R, Bansal D, Hrstka RF (1999) Williamson R. Disruption of the sarcoglycan–sarcospan complex in vascular smooth muscle: a novel mechanism for cardiomyopathy and muscular dystrophy. Cell 98(4):465–74. https://doi.org/10.1016/s0092-8674(00)81975-3
Homburger F, Baker JR, Nixon CW, Whitney R (1962) Primary, generalized polymyopathy and cardiac necrosis in an inbred line of Syrian hamsters. Pharmacology 6(5):339–345
Sakamoto A, Ono K, Abe M, Jasmin G, Eki T, Murakami Y, et al. (1997) Both hypertrophic and dilated cardiomyopathies are caused by mutation of the same gene, delta-sarcoglycan, in hamster: an animal model of disrupted dystrophin-associated glycoprotein complex. Proc Natl Acad Sci USA.94:13873–8. https://doi.org/10.1073/pnas.94.25.13873
Sakamoto A (2003) Molecular pathogenesis of severe cardiomyopathy in the TO-2 hamster. Exp Clin Cardiol 8(3):143
Maekawa K, Hirayama A, Iwata Y, Tajima Y, Nishimaki-Mogami T, Sugawara S, Ueno N, Abe H, Ishikawa M, Murayama M, Matsuzawa Y (2013) Global metabolomic analysis of heart tissue in a hamster model for dilated cardiomyopathy. J Mol Cell Cardiol. 59:76–85. https://doi.org/10.1016/j.yjmcc.2013.02.008
Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA (2006) Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci. 103(5):1394–9. https://doi.org/10.1073/pnas.0507359103
Allikian MJ, Bhabha G, Dospoy P, Heydemann A, Ryder P, Earley JU, Wolf MJ, Rockman HA, McNally EM (2007) Reduced life span with heart and muscle dysfunction in Drosophila sarcoglycan mutants. Hum Mol Genet. 16(23):2933–43. https://doi.org/10.1093/hmg/ddm254
Goldstein JA, Kelly SM, LoPresti PP, Heydemann A, Earley JU, Ferguson EL, Wolf MJ, McNally EM (2011) SMAD signaling drives heart and muscle dysfunction in a Drosophila model of muscular dystrophy. Hum Mol Genet. 20(5):894–904. https://doi.org/10.1093/hmg/ddq528
Wolf MJ (2012) Modeling dilated cardiomyopathies in Drosophila. Trends in cardiovascular medicine. 22(3):55–61. https://doi.org/10.1016/j.tcm.2012.06.012
Ocorr K, Vogler G, Bodmer R (2014) Methods to assess Drosophila heart development, function and aging. Methods 68(1):265–272. https://doi.org/10.1016/j.ymeth.2014.03.031
Walls SM, Cammarato A, Chatfield DA, Ocorr K, Harris GL, Bodmer R (2018) Ceramide-protein interactions modulate ceramide-associated lipotoxic cardiomyopathy. Cell Rep 22(10):2702–2715. https://doi.org/10.1016/j.celrep.2018.02.034
Cammarato A, Dambacher CM, Knowles AF, Kronert WA, Bodmer R, Ocorr K, Bernstein SI (2008) Myosin transducer mutations differentially affect motor function, myofibril structure, and the performance of skeletal and cardiac muscles. Mol Biol Cell 19(2):553–562. https://doi.org/10.1091/mbc.e07-09-0890
Auxerre-Plantié E, Nielsen T, Grunert M, Olejniczak O, Perrot A, Özcelik C, Harries D, Matinmehr F, Dos Remedios C, Mühlfeld C, Kraft T (2020) Identification of MYOM2 as a candidate gene in hypertrophic cardiomyopathy and Tetralogy of Fallot, and its functional evaluation in the Drosophila heart. Disease models & mechanisms. 13(12).
Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, Murata M, Elmén L, Gupta V, Arora S, Sarangi R (2010) A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell 141(1):142–153. https://doi.org/10.1016/j.cell.2010.02.023
Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, Haghighi A, Homsy J, Hubner N, Church G, Cook SA, Linke WA, Chen CS, Seidman JG, Seidman CE (2015)Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy.Science 349:982–986. https://doi.org/10.1126/science.aaa5458
Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC (2012) Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy.Sci Transl Med 4:130ra47. https://doi.org/10.1126/scitranslmed.3003552
Siu CW, Lee YK, Ho JC, Lai WH, Chan YC, Ng KM, Wong LY, Au KW, Lau YM, Zhang J, Lay KW, Colman A, Tse HF (2012) Modeling of lamin A/C mutation premature cardiac aging using patient–specific induced pluripotent stem cells. Aging (Albany NY)4:803–822. https://doi.org/10.18632/aging.100503
Tse HF, Ho JC, Choi SW, Lee YK, Butler AW, Ng KM, Siu CW, Simpson MA, Lai WH, Chan YC, Au KW (2013) Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Human molecular genetics 22(7):1395–403.https://doi.org/10.1093/hmg/dds556
Streckfuss-Bömeke K, Tiburcy M, Fomin A, Luo X, Li W, Fischer C, Özcelik C, Perrot A, Sossalla S, Haas J, Vidal RO (2017) Severe DCM phenotype of patient harboring RBM20 mutation S635A can be modeled by patient-specific induced pluripotent stem cell-derived cardiomyocytes. J Mol Cell Cardiol. 113:9–21.https://doi.org/10.1016/j.yjmcc.2017.09.008
Garg P, Oikonomopoulos A, Chen H, Li Y, Lam CK, Sallam K, Perez M, Lux RL, Sanguinetti MC, Wu JC (2018) Genome editing of induced pluripotent stem cells to decipher cardiac channelopathy variant. J Am Coll Cardiol 72(1):62–75. https://doi.org/10.1016/j.jacc.2018.04.041
Ma N, Zhang JZ, Itzhaki I, Zhang SL, Chen H, Haddad F, Kitani T, Wilson KD, Tian L, Shrestha R, Wu H (2018) Determining the pathogenicity of a genomic variant of uncertain significance using CRISPR/Cas9 and human-induced pluripotent stem cells. Circulation 138(23):2666–81. https://doi.org/10.1161/CIRCULATIONAHA.117.032273
Paik DT, Chandy M, Wu JC (2020) Patient and disease–specific induced pluripotent stem cells for discovery of personalized cardiovascular drugs and therapeutics. Pharmacol Rev. 72(1):320–42. https://doi.org/10.1124/pr.116.013003
Kayvanpour E, Sedaghat-Hamedani F, Amr A, Lai A, Haas J, Holzer DB, Frese KS, Keller A, Jensen K, Katus HA, Meder B (2017) Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clinic Res Cardiol. 106(2):127–39. https://doi.org/10.1007/s00392-016-1033-6
Bondue A, Arbustini E, Bianco A, Ciccarelli M, Dawson D, De Rosa M, Hamdani N, Hilfiker-Kleiner D, Meder B, Leite-Moreira AF, Thum T (2018) Complex roads from genotype to phenotype in dilated cardiomyopathy: scientific update from the Working Group of Myocardial Function of the European Society of Cardiology. Cardiovasc Res. 114(10):1287–303. https://doi.org/10.1093/cvr/cvy122
Houle D, Govindaraju DR, Omholt S (2010) Phenomics: the next challenge. Nat Rev Genet. 11(12):855–66. https://doi.org/10.1038/nrg2897
Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE (2011) HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace 13(8):1077–109. https://doi.org/10.1016/j.hrthm.2011.05.020
Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MR, Towbin JA (2009) Genetic evaluation of cardiomyopathy a Heart Failure Society of America practice guideline. J Card Fail 15(2):83–97.https://doi.org/10.1016/j.cardfail.2018.03.004
Charron P, Arad M, Arbustini E, Basso C, Bilinska Z, Elliott P, Helio T, Keren A, McKenna WJ, Monserrat L, Pankuweit S (2010) Genetic counselling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 31(22):2715–2726. https://doi.org/10.1093/eurheartj/ehq271
Sweet ME, Taylor MR, Mestroni L (2015) Diagnosis, prevalence, and screening of familial dilated cardiomyopathy. Expert opinion on orphan drugs 3(8):869–76. https://doi.org/10.1517/21678707.2015.1057498
Mazzarotto F, Tayal U, Buchan RJ, Midwinter W, Wilk A, Whiffin N, Govind R, Mazaika E, De Marvao A, Dawes TJ, Felkin LE (2020) Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation 141(5):387–98. https://doi.org/10.1161/CIRCULATIONAHA.119.037661
Meune C, Van Berlo JH, Anselme F, Bonne G, Pinto YM, Duboc D (2006) Primary prevention of sudden death in patients with lamin A/C gene mutations. N Engl J Med. 354(2):209–10. https://doi.org/10.1056/NEJMc052632
BÉCANE HM, Bonne G, Varnous S, Muchir A, Ortega V, Hammouda EH, URTIZBEREA JA, Lavergne T, Fardeau M, Eymard B, Weber S (2000) High incidence of sudden death with conduction system and myocardial disease due to lamins A and C gene mutation. Pacing Clin Electrophysiol. 23(11):1661–6. https://doi.org/10.1046/j.1460-9592.2000.01661.x
Chen F, Yang J, Li Y, Wang H (2017) Circulating microRNAs as novel biomarkers for heart failure. Hellenic J Cardiol 59(4):209–14. https://doi.org/10.1016/j.hjc.2017.10.002
Fan KL, Zhang HF, Shen J, Zhang Q, Li XL (2013) Circulating microRNAs levels in Chinese heart failure patients caused by dilated cardiomyopathy. Indian Heart J. 65(1):12–6. https://doi.org/10.1016/j.ihj.2012.12.022
Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K (2007) MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131(1):146–59. https://doi.org/10.1016/j.cell.2007.07.021
Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q (2010) Secreted monocytic miR-150 enhances targeted endothelial cell migration. Molecular cell 39(1):133–44. https://doi.org/10.1016/j.molcel.2010.06.010
Bozkurt B, Colvin M, Cook J, Cooper LT, Deswal A, Fonarow GC, Francis GS, Lenihan D, Lewis EF, McNamara DM, Pahl E (2016). Current diagnostic and treatment strategies for specific dilated cardiomyopathies: a scientific statement from the American Heart Association. Circulation 134(23):e579–646. https://doi.org/10.1161/CIR.0000000000000455
Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE Jr, Drazner MH, Fonarow GC, Geraci SA, Horwich T, Januzzi JL, Johnson MR (2013) ACCF/AHA guideline for the management of heart failure: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guidelines. Circulation 128(16):1810–1852. https://doi.org/10.1161/CIR.0b013e31829e8807
Jessup M, Abraham WT, Casey DE, Feldman AM, Francis GS, Ganiats TG, Konstam MA, Mancini DM, Rahko PS, Silver MA (2009) ACCF/AHA guidelines for the diagnosis and management of heart failure in adults: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation 119(14):1977–2016. https://doi.org/10.1161/CIRCULATIONAHA.109.192064
Dickstein K, Cohen-Solal A, Filippatos G, McMurray JJ, Ponikowski P, Poole-Wilson PA, Strömberg A, vanVeldhuisen DJ, Atar D, Hoes AW (2008) ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure 2008: the Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure 2008 of the European Society of Cardiology. Developed in collaboration with the Heart Failure Association of the ESC (HFA) and endorsed by the Eur Soc Intensive Care Med. (ESICM). Eur Heart J. 29(19):2388–442. https://doi.org/10.1016/j.ejheart.2008.08.005
Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, Morales A, Taylor MR, Vatta M, Ware SM (2018) Genetic evaluation of cardiomyopathy: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med 20(9):899–909. https://doi.org/10.1038/s41436-018-0039-z
Rosenbaum AN, Agre KE, Pereira NL (2020) Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev cardiol. 17(5):286–97. https://doi.org/10.1038/s41569-019-0284-0
Wilcox JE, Hershberger RE (2018) Genetic cardiomyopathies. Current opinion in cardiology 33(3):354–62. https://doi.org/10.1097/HCO.0000000000000512
Funding
Authors have no relevant financial or non-financial interests to disclose.
Author information
Authors and Affiliations
Contributions
Prerna Giri performed the literature survey and prepared the original draft including all figures and table. Dr. Bhagyalaxmi Mohapatra sketched the outline for the article, critically supervised and revised the manuscript at every single step. Amrita Mukhopadhyay prepared and formatted the reference list and collected literature related to next-generation sequencing. Mohini Gupta did formatting of the in-text citations and collected literature on mitochondrial proteins. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Giri, P., Mukhopadhyay, A., Gupta, M. et al. Dilated cardiomyopathy: a new insight into the rare but common cause of heart failure. Heart Fail Rev 27, 431–454 (2022). https://doi.org/10.1007/s10741-021-10125-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10741-021-10125-6