Abstract
Purpose
Invasion of carcinoma cells into surrounding tissue affects breast cancer staging, influences choice of treatment, and impacts on patient outcome. KIF21A is a member of the kinesin superfamily that has been well-studied in congenital extraocular muscle fibrosis. However, its biological relevance in breast cancer is unknown. This study investigated the functional roles of KIF21A in this malignancy and examined its expression pattern in breast cancer tissue.
Methods
The function of KIF21A in breast carcinoma was studied in vitro by silencing its expression in breast cancer cells and examining the changes in cellular activities. Immunohistochemical staining of breast cancer tissue microarrays was performed to determine the expression patterns of KIF21A.
Results
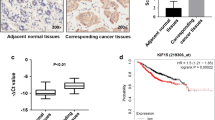
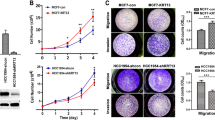
Knocking down the expression of KIF21A using siRNA in MDA-MB-231 and MCF7 human breast cancer cells resulted in significant decreases in tumor cell migration and invasiveness. This was associated with reduced Patched 1 expression and F-actin microfilaments. Additionally, the number of focal adhesion kinase- and paxillin-associated focal adhesions was increased. Immunohistochemical staining of breast cancer tissue microarrays showed that KIF21A was expressed in both the cytoplasmic and nuclear compartments of carcinoma cells. Predominance of cytoplasmic KIF21A was significantly associated with larger tumors and high grade cancer, and prognostic of cause-specific overall patient survival and breast cancer recurrence.
Conclusion
The data demonstrates that KIF21A is an important regulator of breast cancer aggressiveness and may be useful in refining prognostication of this malignant disease.






Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, Znaor A, Bray F (2019) Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 144:1941–1953
Koo CY, Sen YP, Bay BH, Yip GW (2008) Targeting heparan sulfate proteoglycans in breast cancer treatment. Recent Pat Anticancer Drug Discov 3:151–158
Mariotto AB, Yabroff KR, Shao Y, Feuer EJ, Brown ML (2011) Projections of the cost of cancer care in the United States: 2010–2020. J Natl Cancer Inst 103:117–128
Yabroff KR, Lund J, Kepka D, Mariotto A (2011) Economic burden of cancer in the United States: estimates, projections, and future research. Cancer Epidemiol Biomarkers Prev 20:2006–2014
Liu X, Gong H, Huang K (2013) Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Sci 104:651–656
Lucanus AJ, Yip GW (2018) Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene 37:833–838
Zou JX, Duan Z, Wang J, Sokolov A, Xu J, Chen CZ, Li JJ, Chen HW (2014) Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res 12:539–549
Feng YM, Gao G, Zhang F, Chen H, Wan YF, Li XQ (2006) Identification of the differentially expressed genes between primary breast cancer and paired lymph node metastasis through combining mRNA differential display and gene microarray. Zhonghua Yi Xue Za Zhi 86:2749–2755
Feng YM, Wan YF, Li XQ, Cao XC, Li X (2006) Expression and clinical significance of KNSL4 in breast cancer. Ai Zheng 25:744–748
Wang J, Ma S, Ma R, Qu X, Liu W, Lv C, Zhao S, Gong Y (2014) KIF2A silencing inhibits the proliferation and migration of breast cancer cells and correlates with unfavorable prognosis in breast cancer. BMC Cancer 14:461
Corson TW, Huang A, Tsao MS, Gallie BL (2005) KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene 24:4741–4753
Desai J, Velo MP, Yamada K, Overman LM, Engle EC (2012) Spatiotemporal expression pattern of KIF21A during normal embryonic development and in congenital fibrosis of the extraocular muscles type 1 (CFEOM1). Gene Expr Patterns 12:180–188
Yamada K, Andrews C, Chan WM, McKeown CA, Magli A, de Berardinis T, Loewenstein A, Lazar M, O’Keefe M, Letson R, London A, Ruttum M, Matsumoto N, Saito N, Morris L, Del Monte M, Johnson RH, Uyama E, Houtman WA, de Vries B, Carlow TJ, Hart BL, Krawiecki N, Shoffner J, Vogel MC, Katowitz J, Goldstein SM, Levin AV, Sener EC, Ozturk BT, Akarsu AN, Brodsky MC, Hanisch F, Cruse RP, Zubcov AA, Robb RM, Roggenkaemper P, Gottlob I, Kowal L, Battu R, Traboulsi EI, Franceschini P, Newlin A, Demer JL, Engle EC (2003) Heterozygous mutations of the kinesin KIF21A in congenital fibrosis of the extraocular muscles type 1 (CFEOM1). Nat Genet 35:318–321
Marszalek JR, Weiner JA, Farlow SJ, Chun J, Goldstein LS (1999) Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J Cell Biol 145:469–479
Yamada K, Chan WM, Andrews C, Bosley TM, Sener EC, Zwaan JT, Mullaney PB, Ozturk BT, Akarsu AN, Sabol LJ, Demer JL, Sullivan TJ, Gottlob I, Roggenkaemper P, Mackey DA, De Uzcategui CE, Uzcategui N, Ben-Zeev B, Traboulsi EI, Magli A, de Berardinis T, Gagliardi V, Awasthi-Patney S, Vogel MC, Rizzo JF 3rd, Engle EC (2004) Identification of KIF21A mutations as a rare cause of congenital fibrosis of the extraocular muscles type 3 (CFEOM3). Invest Ophthalmol Vis Sci 45:2218–2223
Sun W, Iijima T, Kano J, Kobayashi H, Li D, Morishita Y, Okubo C, Anami Y, Noguchi M (2008) Frequent aberrant methylation of the promoter region of sterile alpha motif domain 14 in pulmonary adenocarcinoma. Cancer Sci 99:2177–2184
Han Q, Han C, Liao X, Huang K, Wang X, Yu T, Yang C, Li G, Han B, Zhu G, Liu Z, Zhou X, Liu J, Su H, Shang L, Peng T, Ye X (2019) Prognostic value of Kinesin-4 family genes mRNA expression in early-stage pancreatic ductal adenocarcinoma patients after pancreaticoduodenectomy. Cancer Med 8:6487–6502
Groth-Pedersen L, Aits S, Corcelle-Termeau E, Petersen NH, Nylandsted J, Jaattela M (2012) Identification of cytoskeleton-associated proteins essential for lysosomal stability and survival of human cancer cells. PLoS One 7:e45381
Iravani O, Bay BH, Yip GW (2017) Silencing HS6ST3 inhibits growth and progression of breast cancer cells through suppressing IGF1R and inducing XAF1. Exp Cell Res 350:380–389
Guo CH, Koo CY, Bay BH, Tan PH, Yip GW (2007) Comparison of the effects of differentially sulphated bovine kidney- and porcine intestine-derived heparan sulphate on breast carcinoma cellular behaviour. Int J Oncol 31:1415–1423
Yu YN, Yip GW, Tan PH, Thike AA, Matsumoto K, Tsujimoto M, Bay BH (2010) Y-box binding protein 1 is up-regulated in proliferative breast cancer and its inhibition deregulates the cell cycle. Int J Oncol 37:483–492
Tan XF, Teo WX, Yip GW (2019) In vitro evaluation of candidate gene targets for cancer therapy. Methods Mol Biol 1974:21–30
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, Zhu J, Haussler D (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38:675–678
Lo SL, Thike AA, Tan SY, Lim TK, Tan IB, Choo SP, Tan PH, Bay BH, Yip GW (2011) Expression of heparan sulfate in gastric carcinoma and its correlation with clinicopathological features and patient survival. J Clin Pathol 64:153–158
Teng YH, Tan PH, Chia SJ, Zam NA, Lau WK, Cheng CW, Bay BH, Yip GW (2008) Increased expression of non-sulfated chondroitin correlates with adverse clinicopathological parameters in prostate cancer. Mod Pathol 21:893–901
Tan PH, Jayabaskar T, Yip G, Tan Y, Hilmy M, Selvarajan S, Bay BH (2005) p53 and c-kit (CD117) protein expression as prognostic indicators in breast phyllodes tumors: a tissue microarray study. Mod Pathol 18:1527–1534
Koo CY, Bay BH, Lui PC, Tse GM, Tan PH, Yip GW (2006) Immunohistochemical expression of heparan sulfate correlates with stromal cell proliferation in breast phyllodes tumors. Mod Pathol 19:1344–1350
Tojkander S, Gateva G, Lappalainen P (2012) Actin stress fibers–assembly, dynamics and biological roles. J Cell Sci 125:1855–1864
Mitra SK, Hanson DA, Schlaepfer DD (2005) Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol 6:56–68
Smilenov LB, Mikhailov A, Pelham RJ, Marcantonio EE, Gundersen GG (1999) Focal adhesion motility revealed in stationary fibroblasts. Science 286:1172–1174
Hooper JE, Scott MP (2005) Communicating with Hedgehogs. Nat Rev Mol Cell Biol 6:306–317
Katoh Y, Katoh M (2004) KIF27 is one of orthologs for Drosophila Costal-2. Int J Oncol 25:1875–1880
Wang ZZ, Yang J, Jiang BH, Di JB, Gao P, Peng L, Su XQ (2018) KIF14 promotes cell proliferation via activation of Akt and is directly targeted by miR-200c in colorectal cancer. Int J Oncol 53:1939–1952
Huang Y, Wang H, Lian Y, Wu X, Zhou L, Wang J, Deng M, Huang Y (2018) Upregulation of kinesin family member 4A enhanced cell proliferation via activation of Akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell Death Dis 9:141
Yip GW, Ferretti P, Copp AJ (2002) Heparan sulphate proteoglycans and spinal neurulation in the mouse embryo. Development 129:2109–2119
Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP (1996) Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev 10:301–312
Edge S, Byrd DR, Compton CC, Fritz AG, Green FL, Trotti A (2010) AJCC cancer staging manual. Springer
Hoda SA, Brogi E, Koerner FC, Rosen PP (2014) Rosen’s breast pathology, 4th edn. Philadelphia, Wolters Kluwer Health
Bouchet BP, Gough RE, Ammon YC, van de Willige D, Post H, Jacquemet G, Altelaar AM, Heck AJ, Goult BT, Akhmanova A (2016) Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. Elife 5.
Weng Z, Shang Y, Yao D, Zhu J, Zhang R (2018) Structural analyses of key features in the KANK1.KIF21A complex yield mechanistic insights into the cross-talk between microtubules and the cell cortex. J Biol Chem 293:215–225
Guo Q, Liao S, Zhu Z, Li Y, Li F, Xu C (2018) Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins. J Biol Chem 293:557–566
Huttenlocher A, Horwitz AR (2011) Integrins in cell migration. Cold Spring Harb Perspect Biol 3:005074
Stehbens SJ, Paszek M, Pemble H, Ettinger A, Gierke S, Wittmann T (2014) CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat Cell Biol 16:561–573
Wozniak MA, Modzelewska K, Kwong L, Keely PJ (2004) Focal adhesion regulation of cell behavior. Biochim Biophys Acta 1692:103–119
Astro V, Chiaretti S, Magistrati E, Fivaz M, de Curtis I (2014) Liprin-alpha1, ERC1 and LL5 define polarized and dynamic structures that are implicated in cell migration. J Cell Sci 127:3862–3876
Li CC, Kuo JC, Waterman CM, Kiyama R, Moss J, Vaughan M (2011) Effects of brefeldin A-inhibited guanine nucleotide-exchange (BIG) 1 and KANK1 proteins on cell polarity and directed migration during wound healing. Proc Natl Acad Sci U S A 108:19228–19233
Kakinuma N, Roy BC, Zhu Y, Wang Y, Kiyama R (2008) Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J Cell Biol 181:537–549
Roy BC, Kakinuma N, Kiyama R (2009) Kank attenuates actin remodeling by preventing interaction between IRSp53 and Rac1. J Cell Biol 184:253–267
Kakinuma N, Kiyama R (2009) A major mutation of KIF21A associated with congenital fibrosis of the extraocular muscles type 1 (CFEOM1) enhances translocation of Kank1 to the membrane. Biochem Biophys Res Commun 386:639–644
Pan W, Sun K, Tang K, Xiao Q, Ma C, Yu C, Wei Z (2018) Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion. J Biol Chem 293:1944–1956
Riobo-Del Galdo NA, Lara Montero A, Wertheimer EV (2019) Role of hedgehog signaling in breast cancer: pathogenesis and therapeutics. Cells 8.
Bhateja P, Cherian M, Majumder S, Ramaswamy B (2019) The Hedgehog signaling pathway: a viable target in breast cancer? Cancers (Basel) 11.
Kubo M, Nakamura M, Tasaki A, Yamanaka N, Nakashima H, Nomura M, Kuroki S, Katano M (2004) Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res 64:6071–6074
Cui W, Wang LH, Wen YY, Song M, Li BL, Chen XL, Xu M, An SX, Zhao J, Lu YY, Mi XY, Wang EH (2010) Expression and regulation mechanisms of Sonic Hedgehog in breast cancer. Cancer Sci 101:927–933
O’Toole SA, Machalek DA, Shearer RF, Millar EK, Nair R, Schofield P, McLeod D, Cooper CL, McNeil CM, McFarland A, Nguyen A, Ormandy CJ, Qiu MR, Rabinovich B, Martelotto LG, Vu D, Hannigan GE, Musgrove EA, Christ D, Sutherland RL, Watkins DN, Swarbrick A (2011) Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res 71:4002–4014
Song L, Wang W, Liu D, Zhao Y, He J, Wang X, Dai Z, Zhang H, Li X (2016) Targeting of sonic hedgehog-Gli signaling: A potential therapeutic target for patients with breast cancer. Oncol Lett 12:1027–1033
Shen T, Ba H, Leng Y, Yan S, Shi J, Yue S, Cheng SY (2019) Sonic Hedgehog stimulates migration of MCF-7 breast cancer cells through Rac1. J Biomed Res 33:297–307
Marigo V, Tabin CJ (1996) Regulation of patched by sonic hedgehog in the developing neural tube. Proc Natl Acad Sci U S A 93:9346–9351
Fabbro M, Henderson BR (2003) Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res 282:59–69
O’Brate A, Giannakakou P (2003) The importance of p53 location: nuclear or cytoplasmic zip code? Drug Resist Updat 6:313–322
Zhang Y, Xiong Y (2001) A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292:1910–1915
Fabbro M, Schuechner S, Au WW, Henderson BR (2004) BARD1 regulates BRCA1 apoptotic function by a mechanism involving nuclear retention. Exp Cell Res 298:661–673
Cmielova J, Rezacova M (2011) p21Cip1/Waf1 protein and its function based on a subcellular localization [corrected]. J Cell Biochem 112:3502–3506
Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E (2010) Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res 704:12–20
Suzuki A, Tsutomi Y, Akahane K, Araki T, Miura M (1998) Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene 17:931–939
Zohny SF, Al-Malki AL, Zamzami MA, Choudhry H (2019) p21(Waf1/Cip1): its paradoxical effect in the regulation of breast cancer. Breast Cancer 26:131–137
Acknowledgements
This project was supported by Grants NMRC/CIRG/1436/2015 and MOH-000152 from the National Medical Research Council, Singapore. A.J.L. was a recipient of the New Colombo Plan Scholarship from the Australian Government’s Department of Foreign Affairs and Trade. X.F.T. and K.W.L. were recipients of the NUSMed Postdoctoral Fellowships from the Yong Loo Lin School of Medicine, National University of Singapore. V.P.C.K. thanks the National University of Singapore for her NUS Research Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Ethics approval 2018/2998 (2011/433/F) for the study was obtained from the SingHealth Centralised Institutional Review Board.
Informed consent
Informed consent was waived for this study as only anonymized analysis was performed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lucanus, A.J., Thike, A.A., Tan, X.F. et al. KIF21A regulates breast cancer aggressiveness and is prognostic of patient survival and tumor recurrence. Breast Cancer Res Treat 191, 63–75 (2022). https://doi.org/10.1007/s10549-021-06426-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-021-06426-x