Abstract
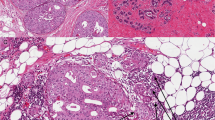
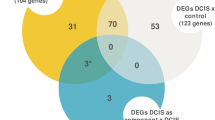
The progression of ductal carcinoma in situ (DCIS) to invasive ductal carcinoma (IDC) marks a critical step in the evolution of breast cancer. There is some evidence to suggest that dynamic interactions between the neoplastic cells and the tumour microenvironment play an important role. Using the whole-genome cDNA-mediated annealing, selection, extension and ligation assay (WG-DASL, Illumina), we performed gene expression profiling on 87 formalin-fixed paraffin-embedded (FFPE) samples from 17 patients consisting of matched IDC, DCIS and three types of stroma: IDC-S (<3 mm from IDC), DCIS-S (<3 mm from DCIS) and breast cancer associated-normal stroma (BC-NS; >10 mm from IDC or DCIS). Differential gene expression analysis was validated by quantitative real time-PCR, immunohistochemistry and immunofluorescence. The expression of several genes was down-regulated in stroma from cancer patients relative to normal stroma from reduction mammoplasties. In contrast, neoplastic epithelium underwent more gene expression changes during progression, including down regulation of SFRP1. In particular, we observed that molecules related to extracellular matrix (ECM) remodelling (e.g. COL11A1, COL5A2 and MMP13) were differentially expressed between DCIS and IDC. COL11A1 was overexpressed in IDC relative to DCIS and was expressed by both the epithelial and stromal compartments but was enriched in invading neoplastic epithelial cells. The contributions of both the epithelial and stromal compartments to the clinically important scenario of progression from DCIS to IDC. Gene expression profiles, we identified differential expression of genes related to ECM remodelling, and specifically the elevated expression of genes such as COL11A1, COL5A2 and MMP13 in epithelial cells of IDC. We propose that these expression changes could be involved in facilitating the transition from in situ disease to invasive cancer and may thus mark a critical point in disease development.





Similar content being viewed by others
Abbreviations
- BC-NS:
-
Breast cancer normal stroma
- DCIS:
-
Ductal carcinoma in situ
- DCIS-S:
-
DCIS-stroma
- ECM:
-
Extracellular matrix
- FF:
-
Fresh frozen
- FFPE:
-
Formalin fixed paraffin embedded
- IDC:
-
Invasive ductal carcinoma
- IDC-S:
-
IDC-stroma
- IF:
-
Immunofluorescence
- IHC:
-
Immunohistochemistry
- ILC:
-
Invasive lobular carcinoma
- LCM:
-
Laser capture microdissection
- NE:
-
Normal epithelium
- qRT-PCR:
-
Quantitative real time-PCR
- RM:
-
Reduction mammoplasty
- RM-NS:
-
Reduction mammoplasty normal stroma
- WG-DASL:
-
Whole-genome cDNA-mediated annealing, selection, extension and ligation assay
References
Wellings SR, Jensen HM (1973) On the origin and progression of ductal carcinoma in the human breast. J Natl Cancer Inst 50(5):1111–1118
Simpson PT, Reis-Filho JS, Gale T, Lakhani SR (2005) Molecular evolution of breast cancer. J Pathol 205(2):248–254
Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchio C, Reis-Filho JS (2010) Breast cancer precursors revisited: molecular features and progression pathways. Histopathology 57(2):171–192
Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC (2003) Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA 100(10):5974–5979
Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC (2009) Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 11(1):R7
Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, Fehm T, Solomayer E, Riess O, Wallwiener D, Kurek R, Neubauer HJ (2006) Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res 66(10):5278–5286
Castro NP, Osorio CA, Torres C, Bastos EP, Mourao-Neto M, Soares FA, Brentani HP, Carraro DM (2008) Evidence that molecular changes in cells occur before morphological alterations during the progression of breast ductal carcinoma. Breast Cancer Res 10(5):R87
Gao Y, Niu Y, Wang X, Wei L, Lu S (2009) Genetic changes at specific stages of breast cancer progression detected by comparative genomic hybridization. J Mol Med 87(2):145–152
Lakhani SR, Collins N, Stratton MR, Sloane JP (1995) Atypical ductal hyperplasia of the breast: clonal proliferation with loss of heterozygosity on chromosomes 16q and 17p. J Clin Pathol 48(7):611–615
Heaphy CM, Bisoffi M, Joste NE, Baumgartner KB, Baumgartner RN, Griffith JK (2009) Genomic instability demonstrates similarity between DCIS and invasive carcinomas. Breast Cancer Res Treat 117:17–24
Knudsen ES, Ertel A, Davicioni E, Kline J, Schwartz GF, Witkiewicz AK (2011) Progression of ductal carcinoma in situ to invasive breast cancer is associated with gene expression programs of EMT and myoepithelia. Breast Cancer Res Treat [Epub ahead of print]
Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D (2011) Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 3(108):108–113
Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14(5):518–527
West RB, Nuyten DS, Subramanian S, Nielsen TO, Corless CL, Rubin BP, Montgomery K, Zhu S, Patel R, Hernandez-Boussard T, Goldblum JR, Brown PO, van de Vijver M, van de Rijn M (2005) Determination of stromal signatures in breast carcinoma. PLoS Biol 3(6):e187
Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO (2004) Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 2(2):E7
Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, Andre S, Piccart M, Campone M, Brain E, Macgrogan G, Petit T, Jassem J, Bibeau F, Blot E, Bogaerts J, Aguet M, Bergh J, Iggo R, Delorenzi M (2009) A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med 15(1):68–74
Waddell N, Cocciardi S, Johnson J, Healey S, Marsh A, Riley J, da Silva L, Vargas AC, Reid L, Simpson PT, Lakhani SR, Chenevix-Trench G (2010) Gene expression profiling of formalin-fixed, paraffin-embedded familial breast tumours using the whole genome-DASL assay. J Pathol 221(4):452–461
April C, Klotzle B, Royce T, Wickham-Garcia E, Boyaniwsky T, Izzo J, Cox D, Jones W, Rubio R, Holton K, Matulonis U, Quackenbush J, Fan JB (2009) Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS ONE 4(12):e8162
Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A (eds) (2010) AJCC cancer staging manual, 7th edn. Springer, New York
Tavassoli FA, Devilee P (2003) World Health Organization Classification of Tumours. Pathology and genetics of tumours of the breast and female genital organs. IARC Press, Lyon
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19(5):403–410
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408
Du P, Kibbe WA, Lin SM (2008) Lumi: a pipeline for processing Illumina microarray. Bioinformatics 24(13):1547–1548
Smyth GK, Michaud J, Scott HS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21(9):2067–2075
Rizki A, Weaver VM, Lee SY, Rozenberg GI, Chin K, Myers CA, Bascom JL, Mott JD, Semeiks JR, Grate LR, Mian IS, Borowsky AD, Jensen RA, Idowu MO, Chen F, Chen DJ, Petersen OW, Gray JW, Bissell MJ (2008) A human breast cell model of preinvasive to invasive transition. Cancer Res 68(5):1378–1387
Delassus GS, Cho H, Eliceiri GL (2011) New signaling pathways from cancer progression modulators to mRNA expression of matrix metalloproteinases in breast cancer cells. J Cell Physiol 226(12):3378–3384
Bowen KB, Reimers AP, Luman S, Kronz JD, Fyffe WE, Oxford JT (2008) Immunohistochemical localization of collagen type XI alpha1 and alpha2 chains in human colon tissue. J Histochem Cytochem 56(3):275–283
Fischer H, Salahshor S, Stenling R, Bjork J, Lindmark G, Iselius L, Rubio C, Lindblom A (2001) COL11A1 in FAP polyps and in sporadic colorectal tumors. BMC Cancer 1:17
Fischer H, Stenling R, Rubio C, Lindblom A (2001) Colorectal carcinogenesis is associated with stromal expression of COL11A1 and COL5A2. Carcinogenesis 22(6):875–878
Halsted KC, Bowen KB, Bond L, Luman SE, Jorcyk CL, Fyffe WE, Kronz JD, Oxford JT (2008) Collagen alpha1(XI) in normal and malignant breast tissue. Mod Pathol 21(10):1246–1254
Nonaka D, Chiriboga L, Rubin BP (2008) Sox10: a pan-schwannian and melanocytic marker. Am J Surg Pathol 32(9):1291–1298
Yang ZQ, Liu G, Bollig-Fischer A, Haddad R, Tarca AL, Ethier SP (2009) Methylation-associated silencing of SFRP1 with an 8p11-12 amplification inhibits canonical and non-canonical WNT pathways in breast cancers. Int J Cancer 125(7):1613–1621
Webb TR, Clark AJ (2010) Minireview: the melanocortin 2 receptor accessory proteins. Mol Endocrinol 24(3):475–484
Saeki T, Tsuruo T, Sato W, Nishikawsa K (2005) Drug resistance in chemotherapy for breast cancer. Cancer Chemother Pharmacol 56(Suppl 1):84–89
Krech T, Scheuerer E, Geffers R, Kreipe H, Lehmann U, Christgen M (2012) ABCB1/MDR1 contributes to the anticancer drug-resistant phenotype of IPH-926 human lobular breast cancer cells. Cancer Lett 315(2):153–160
Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, Gaetje R, Solbach C, Ahr A, Metzler D, Schmidt M, Muller V, Holtrich U, Kaufmann M (2011) A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res 13(5):R97
Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ (2006) Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res 8(5):R61
Kibriya MG, Jasmine F, Roy S, Paul-Brutus RM, Argos M, Ahsan H (2010) Analyses and interpretation of whole-genome gene expression from formalin-fixed paraffin-embedded tissue: an illustration with breast cancer tissues. BMC Genomics 11:622
Mittempergher L, de Ronde JJ, Nieuwland M, Kerkhoven RM, Simon I, Rutgers EJ, Wessels LF, Van’t Veer LJ (2011) Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PLoS ONE 6(2):e17163
Ton CC, Vartanian N, Chai X, Lin MG, Yuan X, Malone KE, Li CI, Dawson A, Sather C, Delrow J, Hsu L, Porter PL (2011) Gene expression array testing of FFPE archival breast tumor samples: an optimized protocol for WG-DASL sample preparation. Breast Cancer Res Treat 125(3):879–883
Reinholz MM, Eckel-Passow JE, Anderson SK, Asmann YW, Zschunke MA, Oberg AL, McCullough AE, Dueck AC, Chen B, April CS, Wickham-Garcia E, Jenkins RB, Cunningham JM, Jen J, Perez EA, Fan JB, Lingle WL (2010) Expression profiling of formalin-fixed paraffin-embedded primary breast tumors using cancer-specific and whole genome gene panels on the DASL(R) platform. BMC Med Genom 3:60
Dahl E, Wiesmann F, Woenckhaus M, Stoehr R, Wild PJ, Veeck J, Knuchel R, Klopocki E, Sauter G, Simon R, Wieland WF, Walter B, Denzinger S, Hartmann A, Hammerschmied CG (2007) Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene 26(38):5680–5691
Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, Mori M, Hirata K, Imai K, Shinomura Y, Baylin SB, Tokino T (2008) Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer 98(6):1147–1156
Ugolini F, Charafe-Jauffret E, Bardou VJ, Geneix J, Adelaide J, Labat-Moleur F, Penault-Llorca F, Longy M, Jacquemier J, Birnbaum D, Pebusque MJ (2001) WNT pathway and mammary carcinogenesis: loss of expression of candidate tumor suppressor gene SFRP1 in most invasive carcinomas except of the medullary type. Oncogene 20(41):5810–5817
Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, Lasorsa L, Letessier A, Ginestier C, Monville F, Esteyries S, Adelaide J, Esterni B, Henry C, Ethier SP, Bibeau F, Mozziconacci MJ, Charafe-Jauffret E, Jacquemier J, Bertucci F, Birnbaum D, Theillet C, Chaffanet M (2005) Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res 3(12):655–667
Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, Rodriguez C, Theillet C (1997) Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res 57(19):4360–4367
Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, Klein J, Fridman E, Skarda J, Srovnal J, Hajduch M, Murray P, Kolar Z (2007) Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer 7:55
Kim H, Watkinson J, Varadan V, Anastassiou D (2010) Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genom 3:51
Acknowledgments
We thank all the patients and their families for the donation of samples for research. We thank Rebecca Johnston and Julie Johnson for excellent technical assistance. Ana Cristina Vargas was supported by a clinical fellowship from the Ludwig Institute for Cancer Research (LICR); Peter T Simpson is the recipient of a Fellowship from the National Breast Cancer Foundation, Australia. Georgia Chenevix-Trench is a Senior Principal Research Fellow of the National Health and Medical Research Council (NHMRC) of Australia. This work was funded in part by grants from the LICR, the NHMRC, the Australian Medical Association Queensland and the Queensland Health Pathology Service—Study, Education and Research Trust Fund.
Conflict of interest
PTS received funding from Illumina to present at a conference about the WG-DASL assay. The remaining authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ana Cristina Vargas and Amy E McCart Reed, and Peter T Simpson and Sunil R Lakhani contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vargas, A.C., Reed, A.E.M., Waddell, N. et al. Gene expression profiling of tumour epithelial and stromal compartments during breast cancer progression. Breast Cancer Res Treat 135, 153–165 (2012). https://doi.org/10.1007/s10549-012-2123-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-012-2123-4