Abstract
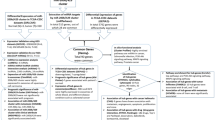
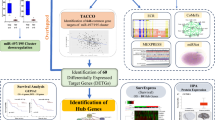
Human papillomavirus accounts for 99.7% of all cervical cancer cases worldwide. The viral oncoproteins alter normal cell signaling and gene expression, resulting in loss of cell cycle control and cancer development. Also, microRNAs (miRNAs) have been reported to play a critical role in cervical carcinogenesis. Especially these are not only appropriate targets for therapeutic intervention in cervical cancer but also early diagnostic signals. The given study tries to improve the sparse knowledge on miRNAs and their role in this physiological context. Deregulated miRNAs were identified by analyzing the raw data of the well-founded GSE20592 dataset including 16 tumor/normal pairs of human cervical tissue samples. The dataset was quantified by a conservative strategy based on HTSeq and Salmon, followed by target prediction via TargetScan and miRDB. The comprehensive pathway analysis of all factors was performed using DAVID. The theoretical results were subject of a stringent experimental validation in a well-characterized clinical cohort of 30 tumor/normal pairs of cervical samples. The top 31 miRNAs and their 140 primary target genes were closely intertwined with the PI3K-Akt signaling pathway. MiR-21-3p and miR-1-3p showed a prominent regulatory role while miR-542, miR-126, miR-143, and miR-26b are directly targeting both PI3K and AKT. This study provides insights into the regulation of PI3K-Akt signaling as an important inducer of cervical cancer and identified miR-542, miR-126, miR-143, and miR-26b as promising inhibitors of the PI3K-Akt action.





Similar content being viewed by others
Availability of Data and Materials
The data that support the findings of this study are available within the paper and Online Resource 1 or on request from the authors.
References
Abdelmohsen K, Srikantan S, Tominaga K et al (2012) Growth inhibition by miR-519 via multiple p21-inducing pathways. Mol Cell Biol 32(13):2530–2548. https://doi.org/10.1128/MCB.00510-12
Abramowitz L, Lacau Saint Guily J, Moyal-Barracco M et al (2018) Epidemiological and economic burden of potentially HPV-related cancers in France. PLoS ONE 13(9):e0202564. https://doi.org/10.1371/journal.pone.0202564
Adil MS, Khulood D, Somanath PR (2021) Targeting Akt-associated microRNAs for cancer therapeutics. BiochemPharmacol 189:114384. https://doi.org/10.1016/j.bcp.2020.114384
Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 31(2):166–169. https://doi.org/10.1093/bioinformatics/btu638
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed Feb 2023
Banelli B, Forlani A, Allemanni G, Morabito A, Pistillo MP, Romani M (2017) MicroRNA in Glioblastoma: an overview. Int J Genomics 2017:7639084. https://doi.org/10.1155/2017/7639084
Banno K, Iida M, Yanokura M et al (2014) MicroRNA in cervical cancer: OncomiRs and Tumor suppressor miRs in diagnosis and treatment. Sci World J 6:178075. https://doi.org/10.1155/2014/178075
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68(6):394–424. https://doi.org/10.3322/caac.21492. [Published correction appears in CA Cancer J Clin. 2020 Jul;70(4):313]
Burmeister CA, Khan SF, Schäfer G et al (2022) Cervical cancer therapies: current challenges and future perspectives. Tumour Virus Res 13:200238. https://doi.org/10.1016/j.tvr.2022.200238
Cai J, Zhao J, Zhang N et al (2015) MicroRNA-542-3p suppresses tumor cell invasion via targeting AKT pathway in human astrocytoma. J Biol Chem 290(41):24678–24688. https://doi.org/10.1074/jbc.M115.649004
Cataldo A, Cheung DG, Balsari A et al (2016) miR-302b enhances breast cancer cell sensitivity to cisplatin by regulating E2F1 and the cellular DNA damage response. Oncotarget. 7(1):786–797. https://doi.org/10.18632/oncotarget.6381
Centomo ML, Vitiello M, Poliseno L, Pandolfi PP (2022) An immunocompetent environment unravels the proto-oncogenic role of miR-22. Cancers (Basel) 14(24):6255. https://doi.org/10.3390/cancers14246255
Chen Y, Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. NucleicAcids Res 48(D1):D127-131. https://doi.org/10.1093/nar/gkz757
Chiantore MV, Mangino G, Iuliano M et al (2016) Human papillomavirus E6 and E7 oncoproteins affect the expression of cancer-related microRNAs: additional evidence in HPV-induced tumorigenesis. J Cancer Res Clin Oncol 142(8):1751–1763. https://doi.org/10.1007/s00432-016-2189-1
Danecek P, Bonfield JK, Liddle J et al (2021) Twelve years of SAMtools and BCFtools. GigaScience. 10(2):giab008. https://doi.org/10.1093/gigascience/giab008
Dixit S, Choi AY, Singh A, Pittala K, Pruett N, Hoang CD (2022) Promising therapeutic potential of tumor suppressor microRNAs for malignant pleural mesothelioma. J Cancer Metastasis Treat 8:47. https://doi.org/10.20517/2394-4722.2022.70
Ferrari E, Gandellini P (2020) Unveiling the ups and downs of miR-205 in physiology and cancer: transcriptional and post-transcriptional mechanisms. Cell Death Dis 11(11):980. https://doi.org/10.1038/s41419-020-03192-4
Fujii T, Shimada K, Asano A et al (2016) MicroRNA-331-3p suppresses cervical cancer cell proliferation and E6/E7 expression by targeting NRP2. Int J Mol Sci 17(8):1351. https://doi.org/10.3390/ijms17081351
Gharbi S, Mohammadi Z, Dezaki MS et al (2022) Characterization of the first microRNA in human CDH1 that affects cell cycle and apoptosis and indicates breast cancers progression. J Cell Biochem 123(3):657–672. https://doi.org/10.1002/jcb.30211
Han Y, Xu GX, Lu H et al (2015) Dysregulation of miRNA-21 and their potential as biomarkers for the diagnosis of cervical cancer. Int J Clin Exp Pathol 8(6):7131–7139
Harden ME, Prasad N, Griffiths A, Munger K (2017) Modulation of microRNA-mRNA target pairs by human papillomavirus 16 oncoproteins. mBio. 8(1):e02170-16. https://doi.org/10.1128/mBio.02170-16
He Y, Sun MM, Zhang GG et al (2021) Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther 6(1):425. https://doi.org/10.1038/s41392-021-00828-5
Hou R, Wang D, Lu J (2017) MicroRNA-10b inhibits proliferation, migration and invasion in cervical cancer cells via direct targeting of insulin-like growth factor-1 receptor. Oncol Lett 13(6):5009–5015. https://doi.org/10.3892/ol.2017.6033
Hu J, Liao D, Sun Z et al (2023) The HPV16 E6, E7/miR-23b-3p/ICAT signaling axis promotes proliferation, migration, invasion and EMT of cervical cancer cells. Carcinogenesis 44(3):221–231. https://doi.org/10.1093/carcin/bgad008
Jiao D, Chen J, Li Y et al (2018) miR-1-3p and miR-206 sensitizes HGF-induced gefitinib-resistant human lung cancer cells through inhibition of c-Met signalling and EMT. J Cell Mol Med 22(7):3526–3536. https://doi.org/10.1111/jcmm.13629
Kakotkin VV, Semina EV, Zadorkina TG et al (2023) Prevention strategies and early diagnosis of cervical cancer: current state and prospects. Diagnostics (Basel). 13(4):610. https://doi.org/10.3390/diagnostics13040610
Kang H, Kim C, Ji E et al (2019) The MicroRNA-551a/MEF2C axis regulates the survival and sphere formation of cancer cells in response to 5-fluorouracil. Mol Cells 42(2):175–182. https://doi.org/10.14348/molcells.2018.0288
Kim YW, Kim EY, Jeon D et al (2014) Differential microRNA expression signatures and cell type-specific association with Taxol resistance in ovarian cancer cells. Drug Des Devel Ther 8:293–314. https://doi.org/10.2147/DDDT.S51969
Krueger F, Trimgalore (2021) GitHub repository. https://github.com/FelixKrueger/TrimGalore. Accessed Feb 2023
Lajer CB, Garnæs E, Friis-Hansen L et al (2012) The role of miRNAs in human papillomavirus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer 106(9):1526–1534. https://doi.org/10.1038/bjc.2012.109
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Lawrence MS, Stojanov P, Mermel CH et al (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505(7484):495–501. https://doi.org/10.1038/nature12912
Lei J, Deng F, Ding H et al (2022) Recent developments on the roles of calcium signals and potential therapy targets in cervical cancer. Cells. 11(19):3003. https://doi.org/10.3390/cells11193003
Li SM, Wu HL, Yu X et al (2018) The putative tumour suppressor miR-1-3p modulates prostate cancer cell aggressiveness by repressing E2F5 and PFTK1. J Exp Clin Cancer Res 37(1):219. https://doi.org/10.1186/s13046-018-0895-z
Liontos M, Kyriazoglou A, Dimitriadis I, Dimopoulos MA, Bamias A (2019) Systemic therapy in cervical cancer: 30 years in review. Crit Rev Oncol Hematol 137:9–17. https://doi.org/10.1016/j.critrevonc.2019.02.009
Liu J, Huang Y, Cheng Q et al (2019) miR-1-3p suppresses the epithelial-mesenchymal transition property in renal cell cancer by downregulating Fibronectin 1. Cancer Manag Res 11:5573–5587. https://doi.org/10.2147/CMAR.S200707
Liu J, Li Y, Chen X et al (2020a) Upregulation of miR-205 induces CHN1 expression, which is associated with the aggressive behaviour of cervical cancer cells and correlated with lymph node metastasis. BMC Cancer 20(1):1029. https://doi.org/10.1186/s12885-020-07478-w
Liu QH, Li HL, Zhang H, Li Z, Zhao M, Zhang TT (2020b) miR-542 inhibits tumorigenesis and chemoresistance through the AKT/NFκB pathway in cervical cancer. J Biol RegulHomeost Agents 34(5):1809–1817. https://doi.org/10.23812/20-265-L
Liu Y, Li L, Yang Z, Wen D, Hu Z (2022) Circular RNA circACAP2 suppresses ferroptosis of cervical cancer during malignant progression by miR-193a-5p/GPX4. J Oncol 2022:5228874. https://doi.org/10.1155/2022/5228874
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Ma J, Zhang F, Sun P (2020) miR-140-3p impedes the proliferation of human cervical cancer cells by targeting RRM2 to induce cell-cycle arrest and early apoptosis. Bioorg Med Chem 28(3):115283. https://doi.org/10.1016/j.bmc.2019.115283
Martínez-Noël G, Szajner P, Kramer RE et al (2022) Identification of MicroRNAs that stabilize p53 in human papillomavirus-positive cancer cells. J Virol 96(4):e0186521. https://doi.org/10.1128/JVI.01865-21
McGeary SE, Lin KS, Shi CY et al (2019) The biochemical basis of microRNA targeting efficacy. Science. 366(6472):eaav1741. https://doi.org/10.1126/science.aav1741
Mellman I, Yarden Y (2013) Endocytosis and cancer. Cold Spring Harb Perspect Biol. 5(12):a016949. https://doi.org/10.1101/cshperspect.a016949
Menges CW, Baglia LA, Lapoint R, McCance DJ (2006) Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res 66(11):5555–5559. https://doi.org/10.1158/0008-5472.CAN-06-0499
Mesri EA, Feitelson MA, Munger K (2014) Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 15:266–282. https://doi.org/10.1016/j.chom.2014.02.011
Mutlu M, Saatci Ö, Ansari SA et al (2016) MiR-564 acts as a dual inhibitor of PI3K and MAPK signaling networks and inhibits proliferation and invasion in breast cancer. Sci Rep 6(1):1–14. https://doi.org/10.1038/srep32541
Nouraee N, Van Roosbroeck K, Vasei M et al (2013) Expression, tissue distribution and function of miR-21 in esophageal squamous cell carcinoma. PLoS ONE 8(9):e73009. https://doi.org/10.1371/journal.pone.0073009
Pan X, Yang W, Wen Z, Li F, Tong L, Tang W (2020) Does adenocarcinoma have a worse prognosis than squamous cell carcinoma in patients with cervical cancer? A real-world study with a propensity score matching analysis. J Gynecol Oncol 31(6):e80. https://doi.org/10.3802/jgo.2020.31.e80
Park S, Eom K, Kim J et al (2017) MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer 17(1):658. https://doi.org/10.1186/s12885-017-3642-5
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14(4):417–419. https://doi.org/10.1038/nmeth.4197
Pim D, Massimi P, Dilworth S, Banks L (2005) Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene 24:7830–7838. https://doi.org/10.1038/sj.onc.1208935
Prati B, Marangoni B, Boccardo E (2018) Human papillomavirus and genome instability: from productive infection to cancer. Clinics (Sao Paulo, Brazil) 73(suppl 1):e539s. https://doi.org/10.6061/clinics/2018/e539s
R Core Team (2023) R: a language and environment for statistical computing, Vienna, Austria. https://www.R-project.org/. Accessed March 2023
Rhim J, Baek W, Seo Y, Kim JH (2022) From molecular mechanisms to therapeutics: understanding MicroRNA-21 in cancer. Cells 11(18):2791. https://doi.org/10.3390/cells11182791
Ruttkay-Nedecky B, Jimenez Jimenez AM, Nejdl L et al (2013) Relevance of infection with human papillomavirus: the role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins. Int J Oncol 43:1754–1762. https://doi.org/10.3892/ijo.2013.2105
Schüller U, Ruiter M, Herms J, Kretzschmar HA, Grasbon-Frodl E (2008) Absence of mutations in the AKT1 oncogene in glioblastomas and medulloblastomas. Acta Neuropathol 115:367–368. https://doi.org/10.1007/s00401-007-0334-2
Sherman BT, Hao M, Qiu J et al (2022) DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 50(W1):W216–W221. https://doi.org/10.1093/nar/gkac194
Skelin J, Sabol I, Tomaić V (2022) Do or Die: HPV E5, E6 and E7 in cell death evasion. Pathogens 11(9):1027. https://doi.org/10.3390/pathogens11091027
Snoek BC, Babion I, Koppers-Lalic D, Pegtel DM, Steenbergen RD (2019) Altered microRNA processing proteins in HPV-induced cancers. Curr Opin Virol 39:23–32. https://doi.org/10.1016/j.coviro.2019.07.002
Sun X, Hou L, Qiu C, Kong B (2021) MiR-501 promotes tumor proliferation and metastasis by targeting HOXD10 in endometrial cancer. Cell Mol Biol Lett 26(1):20. https://doi.org/10.1186/s11658-021-00268-7
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Tan D, Zhou C, Han S, Hou X, Kang S, Zhang Y (2018) MicroRNA-378 enhances migration and invasion in cervical cancer by directly targeting autophagy-related protein 12. Mol Med Rep 17(5):6319–6326. https://doi.org/10.3892/mmr.2018.8645
Venuti A, Paolini F, Nasir L et al (2011) Papillomavirus E5: the smallest oncoprotein with many functions. Mol Cancer 10:140. https://doi.org/10.1186/1476-4598-10-140
Wang JY, Huang JC, Chen G, Wei DM (2018) Expression level and potential target pathways of miR-1-3p in colorectal carcinoma based on 645 cases from 9 microarray datasets. Mol Med Rep 17(4):5013–5020. https://doi.org/10.3892/mmr.2018.8532
Wang H, Hu H, Luo Z et al (2020) miR-4454 up-regulated by HPV16 E6/E7 promotes invasion and migration by targeting ABHD2/NUDT21 in cervical cancer. Biosci Rep. 40(9):BSR20200796. https://doi.org/10.1042/BSR20200796
Wang T, Zhang XD, Hua KQ (2021) A ceRNA network of BBOX1-AS1-hsa-miR-125b-5p/hsa-miR-125a-5p-CDKN2A shows prognostic value in cervical cancer. Taiwan J Obstet Gynecol 60(2):253–261. https://doi.org/10.1016/j.tjog.2020.12.006
World Health Organization (2014) Comprehensive Cervical Cancer Control: A Guide to Essential Practice. 2nd ed. World Health Organization.
WHO. Cervical cancer 2024. [Accessed on March 2024]. Available at: http://www.who.int/mediacentre/factsheets/fs380/en/
WHO. Human papillomavirus vaccines (HPV) 2024. [Accessed on March 2024]. Available at: https://www.who.int/teams/immunization-vaccines-and-biologicals/diseases/human-papillomavirus-vaccines-(HPV)
Witten D, Tibshirani R, Gu SG, Fire A, Lui WO (2010) Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol. https://doi.org/10.1186/1741-7007-8-58
Xie H, Jing R, Liao X et al (2022) Arecoline promotes proliferation and migration of human HepG2 cells through activation of the PI3K/AKT/mTOR pathway. Hereditas 159(1):29. https://doi.org/10.1186/s41065-022-00241-0
Xu M, Sun J, Yu Y et al (2020) TM4SF1 involves in miR-1-3p/miR-214-5p-mediated inhibition of the migration and proliferation in keloid by regulating AKT/ERK signaling. Life Sci 254:117746. https://doi.org/10.1016/j.lfs.2020.117746
Xu W, Wang Z, Zhang Z, Xu J, Jiang Y (2022) PIK3CB promotes oesophageal cancer proliferation through the PI3K/AKT/mTOR signalling axis. Cell Biol Int 46(9):1399–1408. https://doi.org/10.1002/cbin.11847
Zhang L, Wu J, Ling MT, Zhao L, Zhao KN (2015) The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol Cancer 14:87. https://doi.org/10.1186/s12943-015-0361-x
Zhang J, Wang L, Mao S et al (2018a) miR-1-3p contributes to cell proliferation and invasion by targeting glutaminase in bladder cancer cells. Cell PhysiolBiochem 51(2):513–527. https://doi.org/10.1159/000495273
Zhang X, Liu Y, Huang WC, Zheng LC (2018b) MiR-125b-1-3p exerts antitumor functions in lung carcinoma cells by targeting S1PR1. Chin Med J (Engl) 131(16):1909–1916. https://doi.org/10.4103/0366-6999.238135
Zhang P, Lu X, Shi Z et al (2019) miR-205-5p regulates epithelial-mesenchymal transition by targeting PTEN via PI3K/AKT signaling pathway in cisplatin-resistant nasopharyngeal carcinoma cells. Gene 710:103–113. https://doi.org/10.1016/j.gene.2019.05.058
Zhao S, Yao D, Chen J, Ding N, Ren F (2015) MiR-20a promotes cervical cancer proliferation and metastasis in vitro and in vivo. PLoS ONE 10(3):e0120905. https://doi.org/10.1371/journal.pone.0120905
Zhu Y, Tang H, Zhang L et al (2019) Suppression of miR-21-3p enhances TRAIL-mediated apoptosis in liver cancer stem cells by suppressing the PI3K/Akt/Bad cascade via regulating PTEN. Cancer Manag Res 11:955–968. https://doi.org/10.2147/CMAR.S183328
Acknowledgements
The authors wish to thank all patients who participated in this study.
Funding
This study was supported by faculty research grants of the Alzahra University.
Author information
Authors and Affiliations
Contributions
Akram Rahimi-Moghaddam: Performed the bioinformatics analyses and experiments and wrote the manuscript. Nassim Ghorbanmehr: Supervised and designed the study, wrote the manuscript, and performed data analysis. Sedigheh Gharbi and Eberhard Korsching: Data analysis and review of the manuscript. Fatemeh Nili: Sample preparation and manuscript review. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no financial or non-financial interests that might influence the results and/or discussion reported in this paper.
Ethical approval and consent to participate
All experiments involving human samples were reviewed and approved by the Ethics Committee of Alzahra University (IR.ALZAHRA.REC.1400.058).
Informed Consent
Informed consent was obtained from all patients prior to the use of the samples for scientific research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rahimi-Moghaddam, A., Ghorbanmehr, N., Gharbi, S. et al. Interplay of miR-542, miR-126, miR-143 and miR-26b with PI3K-Akt is a Diagnostic Signal and Putative Regulatory Target in HPV-Positive Cervical Cancer. Biochem Genet (2024). https://doi.org/10.1007/s10528-024-10837-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10528-024-10837-y