Abstract
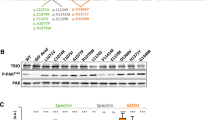
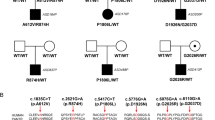
ASH1L potentially contributes to Tourette syndrome (TS) and other neuropsychiatric disorders, as our previous studies have shown. It regulates essential developmental genes by counteracting polycomb-mediated transcriptional repression, which restricts chromatin accessibility at target genes. ASH1L is highly expressed in the adult brain, playing a crucial role in the early stage. However, it remains unclear how ASH1L mutations carried by patients with TS participate in regulating neuronal growth processes leading to TS traits. Five TS families recruited in our study underwent comprehensive physical examinations and questionnaires to record clinical phenotypes and environmental impact factors. We validated the variants via Sanger sequencing and constructed two mutants near the catalytic domain of ASH1L. We conducted molecular modeling, in vitro assays, and primary neuron cultures to find the role of ASH1L in neuronal development and its correlation with TS. In this study, we validated five pathogenic ASH1L rare variants and observed symptoms in patients with simple tics and behavioral comorbidities. Mutations near the catalytic domain of TS patients cause mental state abnormalities and disrupt ASH1L function by destabilizing its spatial conformation, leading to decreased activity of catalytic H3K4, thereby affecting the neurite growth. We need to conduct larger-scale studies on TS patients and perform additional neurological evaluations on mature neurons. We first reported the effects of ASH1L mutations in TS patients, including phenotypic heterogeneity, protein function, and neurological growth. This information contributes to understanding the neurodevelopmental pathogenesis of TS in patients with ASH1L mutations.




Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data availability
The datasets used and analyzed in the current study are available from the corresponding author upon reasonable request.
References
Hartmann A, Worbe Y (2018) Tourette syndrome: clinical spectrum, mechanisms and personalized treatments. Curr Opin Neurol 31(4):504–509. https://doi.org/10.1097/WCO.0000000000000575
Johnson KA, Worbe Y, Foote KD et al (2023) Tourette syndrome: clinical features, pathophysiology, and treatment. Lancet Neurol 22(2):147–158. https://doi.org/10.1016/S1474-4422(22)00303-9
Deeb W, Malaty IA, Mathews CA (2019) Tourette disorder and other tic disorders. Handb Clin Neurol 165:123–153. https://doi.org/10.1016/B978-0-444-64012-3.00008-3
Lin WD, Tsai FJ, Chou IC (2022) Current understanding of the genetics of Tourette syndrome. Biomed J 45(2):271–279. https://doi.org/10.1016/j.bj.2022.01.008
Browne HA, Hansen SN, Buxbaum JD et al (2015) Familial clustering of tic disorders and obsessive-compulsive disorder. JAMA Psychiat 72(4):359–366. https://doi.org/10.1001/jamapsychiatry.2014.2656
Vadgama N, Pittman A, Simpson M et al (2019) De novo single-nucleotide and copy number variation in discordant monozygotic twins reveals disease-related genes. Eur J Hum Genet 27(7):1121–1133. https://doi.org/10.1038/s41431-019-0376-7
Zhang C, Xu L, Zheng X, Liu S, Che F (2021) Role of Ash1l in Tourette syndrome and other neurodevelopmental disorders. Dev Neurobiol 81(2):79–91. https://doi.org/10.1002/dneu.22795
Liu S, Tian M, He F et al (2020) Mutations in ASH1L confer susceptibility to Tourette syndrome. Mol Psychiatry 25(2):476–490. https://doi.org/10.1038/s41380-019-0560-8
Liu W, Xu L, Zhang C et al (2022) ASH1L may contribute to the risk of Tourette syndrome: combination of family-based analysis and case-control study. Brain Behav 12(4):e2539. https://doi.org/10.1002/brb3.2539
Dorighi KM, Tamkun JW (2013) The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development 140(20):4182–4192. https://doi.org/10.1242/dev.095786
Widiger TA, Hines A (2022) The diagnostic and statistical manual of mental disorders, fifth edition alternative model of personality disorder. Personal Disord 13(4):347–355. https://doi.org/10.1037/per0000524
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Yan Y, Tian M, Li M et al (2022) ASH1L haploinsufficiency results in autistic-like phenotypes in mice and links Eph receptor gene to autism spectrum disorder. Neuron 110(7):1156-1172.e9. https://doi.org/10.1016/j.neuron.2021.12.035
Liu S, Zheng L, Zheng X et al (2017) The subjective quality of life in young people with Tourette syndrome in China. J Atten Disord 21(5):426–432. https://doi.org/10.1177/1087054713518822
Roessner V, Eichele H, Stern JS et al (2022) European clinical guidelines for Tourette syndrome and other tic disorders-version 2.0. Part III: pharmacological treatment. Eur Child Adolesc Psychiatry 31(3):425–441. https://doi.org/10.1007/s00787-021-01899-z
Okamoto N, Miya F, Tsunoda T et al (2017) Novel MCA/ID syndrome with ASH1L mutation. Am J Med Genet A 173(6):1644–1648. https://doi.org/10.1002/ajmg.a.38193
Shen W, Krautscheid P, Rutz AM, Bayrak-Toydemir P, Dugan SL (2019) De novo loss-of-function variants of ASH1L are associated with an emergent neurodevelopmental disorder. Eur J Med Genet 62(1):55–60. https://doi.org/10.1016/j.ejmg.2018.05.003
Nakamura T, Blechman J, Tada S et al (2000) huASH1 protein, a putative transcription factor encoded by a human homologue of the Drosophila ash1 gene, localizes to both nuclei and cell-cell tight junctions. Proc Natl Acad Sci U S A 97(13):7284–7289. https://doi.org/10.1073/pnas.97.13.7284
Yuan W, Xu M, Huang C et al (2011) H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286(10):7983–7989. https://doi.org/10.1074/jbc.M110.194027.H3K36
Wysocka J, Swigut T, Xiao H et al (2006) A PhD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442(7098):86–90. https://doi.org/10.1038/nature04815
An S, Yeo KJ, Jeon YH, Song JJ (2011) Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J Biol Chem 286(10):8369–8374. https://doi.org/10.1074/jbc.M110.203380
Rogawski DS, Ndoj J, Cho HJ et al (2015) Two loops undergoing concerted dynamics regulate the activity of the ASH1L histone methyltransferase. Biochemistry 54(35):5401–5413. https://doi.org/10.1021/acs.biochem.5b00697
Lee Y, Yoon E, Cho S et al (2019) Structural basis of MRG15-mediated activation of the ASH1L histone methyltransferase by releasing an autoinhibitory loop. Structure 27(5):846-852.e3. https://doi.org/10.1016/j.str.2019.01.016
Yu M, Jia Y, Ma Z et al (2022) Structural insight into ASH1L PhD finger recognizing methylated histone H3K4 and promoting cell growth in prostate cancer. Front Oncol 12:906807. https://doi.org/10.3389/fonc.2022.906807
Faundes V, Newman WG, Bernardini L et al (2018) Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am J Hum Genet 102(1):175–187. https://doi.org/10.1016/j.ajhg.2017.11.013
Xia M, Liu J, Wu X et al (2013) Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity 39(3):470–481. https://doi.org/10.1016/j.immuni.2013.08.016
Zhao F, Liu Y, Su X et al (2020) Molecular basis for histone H3 “K4me3-K9me3/2” methylation pattern readout by Spindlin1. J Biol Chem 295(49):16877–16887. https://doi.org/10.1074/jbc.RA120.013649
Corley M, Kroll KL (2015) The roles and regulation of Polycomb complexes in neural development. Cell Tissue Res 359(1):65–85. https://doi.org/10.1007/s00441-014-2011-9
Boyer LA, Plath K, Zeitlinger J et al (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441(7091):349–353. https://doi.org/10.1038/nature04733
Acknowledgements
We thank all the participants in our research and the contributions of all investigators.
Funding
Grants from the National Natural Science Foundation (NSFC) (No. 81371499) and the National Key Research and Development Program of China (Nos. 2016YFC1000306 and 2005DKA32408) supported this work.
Author information
Authors and Affiliations
Contributions
Data curation, C.Z. and W.L.; Formal analysis, C.Z. and L.X.; Funding acquisition, S.L. and F.C.; Investigation, C.Z. and W.L.; Project administration, S.L. and F.C.; Resources, C.Z., W.L., L.X., S.L. and F.C.; Supervision, S.L. and F.C.; Validation, C.Z. and L.X.; Writing–original draft, C.Z.; Writing–review & editing, C.Z., S.L. and F.C. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, C., Liu, W., Xu, L. et al. Abnormal H3K4 enzyme catalytic activity and neuronal morphology caused by ASH1L mutations in individuals with Tourette syndrome. Eur Child Adolesc Psychiatry 33, 3913–3923 (2024). https://doi.org/10.1007/s00787-024-02437-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00787-024-02437-3