Abstract
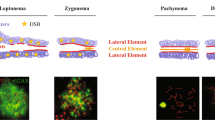
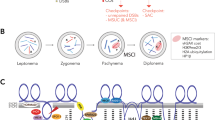
Eutherian mammals have an extremely conserved sex-determining system controlled by highly differentiated sex chromosomes. Females are XX and males XY, and any deviation generally leads to infertility, mainly due to meiosis disruption. The African pygmy mouse (Mus minutoides) presents an atypical sex determination system with three sex chromosomes: the classical X and Y chromosomes and a feminizing X chromosome variant, called X*. Thus, three types of females coexist (XX, XX*, and X*Y) that all show normal fertility. Moreover, the three chromosomes (X and Y on one side and X* on the other side) are fused to different autosomes, which results in the inclusion of the sex chromosomes in a quadrivalent in XX* and X*Y females at meiotic prophase. Here, we characterized the configurations adopted by these sex chromosome quadrivalents during meiotic prophase. The XX* quadrivalent displayed a closed structure in which all homologous chromosome arms were fully synapsed and with sufficient crossovers to ensure the reductional segregation of all chromosomes at the first meiotic division. Conversely, the X*Y quadrivalents adopted either a closed configuration with non-homologous synapsis of the X* and Y chromosomes or an open chain configuration in which X* and Y remained asynapsed and possibly transcriptionally silenced. Moreover, the number of crossovers was insufficient to ensure chromosome segregation in a significant fraction of nuclei. Together, these findings raise questions about the mechanisms allowing X*Y females to have a level of fertility as good as that of XX and XX* females, if not higher.





Similar content being viewed by others
References
Akhverdyan M, Fredga K (2001) EM studies of female meiosis in wood lemmings with different sex chromosome constitutions. J Exp Zool 290:504–516
Alton M, Lau MP, Villemure M, Taketo T (2008) The behavior of the X- and Y-chromosomes in the oocyte during meiotic prophase in the B6.YTIRsex-reversed mouse ovary. Reproduction. 135:241–252
Amleh A, Smith L, Chen H-Y, Taketo T (2000) Both nuclear and cytoplasmic components are defective in oocytes of the B6.YTIR sex-reversed female mouse. Dev Biol 219:277–286. https://doi.org/10.1006/dbio.1999.9600
Arboleda VA, Fleming A, Barseghyan H, Délot E, Sinsheimer JS, Vilain E (2014) Regulation of sex determination in mice by a non-coding genomic region. Genetics 197:885–897. https://doi.org/10.1534/genetics.113.160259
Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JHJ, de Boer P, Grootegoed JA (2005) Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol 25:1041–1053
Bachtrog D (2013) Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14:113–124. https://doi.org/10.1038/nrg3366
Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice W, Valenzuela N (2011) Are all sex chromosomes created equal? Trends Genet 27:350–357. https://doi.org/10.1016/j.tig.2011.05.005
Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13:336–342
Baudat F, Imai Y, de Massy B (2013) Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14:794–806. https://doi.org/10.1038/nrg3573
Borodin PM, Basheva EA, Torgasheva AA, Dashkevich OA, Golenishchev FN, Kartavtseva IV, Mekada K, Dumont BL (2012) Multiple independent evolutionary losses of XY pairing at meiosis in the grey voles. Chromosom Res 20:259–268. https://doi.org/10.1007/s10577-011-9261-0
Britton-Davidian J, Robinson TJ, Veyrunes F (2012) Systematics and evolution of the African pygmy mice, subgenus Nannomys: a review. Acta Oecol 42:41–49. https://doi.org/10.1016/j.actao.2012.01.001
Burgoyne PS, Baker TG (1981) Oocyte depletion in XO mice and their XX sibs from 12 to 200 days post partum. J Reprod Fertil 61:207–212
Burgoyne PS, Baker TG (1985) Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. J Reprod Fertil 75:633–645
Burgoyne PS, Mahadevaiah SK, Turner JM (2009) The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 10:207–216
Casey AE, Daish TJ, Barbero JL, Grützner F (2017) Differential cohesin loading marks paired and unpaired regions of platypus sex chromosomes at prophase I. Sci Rep 7:4217. https://doi.org/10.1038/s41598-017-04560-5
Charlesworth D, Charlesworth B, Marais G (2005) Steps in the evolution of heteromorphic sex chromosomes. Heredity (Edinb) 95:118–128. https://doi.org/10.1038/sj.hdy.6800697
Cloutier JM, Mahadevaiah SK, ElInati E, Nussenzweig A, Tóth A, Turner JMA (2015) Histone H2AFX links meiotic chromosome asynapsis to prophase I oocyte loss in mammals. PLoS Genet 11:e1005462. https://doi.org/10.1371/journal.pgen.1005462
Cloutier JM, Mahadevaiah SK, ElInati E, Tóth A, Turner J (2016) Mammalian meiotic silencing exhibits sexually dimorphic features. Chromosoma 125:215–226. https://doi.org/10.1007/s00412-015-0568-z
Daish T, Casey A, Grützner F (2009) Platypus chain reaction: directional and ordered meiotic pairing of the multiple sex chromosome chain in Ornithorhynchus anatinus. Reprod Fertil Dev 21:976–984. https://doi.org/10.1071/RD09085
de la Fuente R, Parra MT, Viera A, Calvente A, Gómez R, Suja JÁ, Rufas JS, Page J (2007) Meiotic pairing and segregation of achiasmate sex chromosomes in eutherian mammals: the role of SYCP3 protein. PLoS Genet 3:e198. https://doi.org/10.1371/journal.pgen.0030198
Deuve JL, Bennett NC, Ruiz-Herrera A, Waters PD, Britton-Davidian J, Robinson TJ (2008) Dissection of a Y-autosome translocation in Cryptomys hottentotus (Rodentia, Bathyergidae) and implications for the evolution of a meiotic sex chromosome chain. Chromosoma 117:211–217. https://doi.org/10.1007/s00412-007-0140-6
Dobigny G, Ozouf-Costaz C, Bonillo C, Volobouev V (2004) Viability of X-autosome translocations in mammals: an epigenomic hypothesis from a rodent case-study. Chromosoma 113:34–41. https://doi.org/10.1007/s00412-004-0292-6
Eicher EM, Washburn LL, Whitney JB, Morrow KE (1982) Mus poschiavinus Y chromosome in the C57BL/6J murine genome causes sex reversal. Science 217:535–537
Fredga K (1970) Unusual sex chromosome inheritance in mammals. Philos Trans R Soc Lond Ser B Biol Sci 259:15–36
Fredga K (1994) Bizarre mammalian sex-determining mechanisms. In: Short R, Balaban E (eds) The differences between the sexes. Cambridge University Press, Cambridge, pp 419–431
Grey C, Baudat F, de Massy B (2009) Genome-wide control of the distribution of meiotic recombination. PLoS Biol 7:e35
Gruetzner F, Ashley T, Rowell DM, Marshall Graves JA (2006) How did the platypus get its sex chromosome chain? A comparison of meiotic multiples and sex chromosomes in plants and animals. Chromosoma 115:75–88. https://doi.org/10.1007/s00412-005-0034-4
Gubbay J, Vivian N, Economou A, Jackson D, Goodfellow P, Lovell-Badge R (1992) Inverted repeat structure of the Sry locus in mice. Proc Natl Acad Sci U S A 89:7953–7957
Guillon H, Baudat F, Grey C, Liskay RM, de Massy B (2005) Crossover and noncrossover pathways in mouse meiosis. Mol Cell 20:563–573. https://doi.org/10.1016/j.molcel.2005.09.021
Huang L, Chi J, Wang J, Nie W, Su W, Yang F (2006) High-density comparative BAC mapping in the black muntjac (Muntiacus crinifrons): molecular cytogenetic dissection of the origin of MCR 1p+4 in the X1X2Y1Y2Y3 sex chromosome system. Genomics 87:608–615. https://doi.org/10.1016/j.ygeno.2005.12.008
Hunter N (2015) Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol 7:a016618. https://doi.org/10.1101/cshperspect.a016618
Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH (2011) MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev 25:959–971. https://doi.org/10.1101/gad.2030811
Jotterand-Bellomo M (1981) Le caryotype et la spermatogénèse de Mus setulosus (bandes Q, C, G et coloration argentique). Genetica 56:217–227. https://doi.org/10.1007/BF00057563
Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S (2013) Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 27:873–886. https://doi.org/10.1101/gad.213652.113
Kolas NK, Marcon E, Crackower MA, Höög C, Penninger JM, Spyropoulos B, Moens PB (2005a) Mutant meiotic chromosome core components in mice can cause apparent sexual dimorphic endpoints at prophase or X-Y defective male-specific sterility. Chromosoma 114:92–102
Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, Greally J, Edelmann W, Cohen PE (2005b) Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol 171:447–458
Lampson MA, Black BE (2017) Cellular and molecular mechanisms of centromere drive. Cold Spring Harb Symp Quant Biol 82:249–257. https://doi.org/10.1101/sqb.2017.82.034298
Laval SH, Glenister PH, Rasberry C, Thornton CE, Mahadevaiah SK, Cooke HJ, Burgoyne PS, Cattanach BM (1995) Y chromosome short arm-Sxr recombination in XSxr/Y males causes deletion of Rbm and XY female sex reversal. Proc Natl Acad Sci U S A 92:10403–10407
LeMaire-Adkins R, Hunt PA (2000) Nonrandom segregation of the mouse univalent X chromosome: evidence of spindle-mediated meiotic drive. Genetics 156:775–783
Mahadevaiah SK, Lovell-Badge R, Burgoyne PS (1993) Tdy-negative XY, XXY and XYY female mice: breeding data and synaptonemal complex analysis. J Reprod Fertil 97:151–160
Marcon E, Moens P (2003) MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165:2283–2287
Matveevsky S, Bakloushinskaya I, Kolomiets O (2016) Unique sex chromosome systems in Ellobius: how do male XX chromosomes recombine and undergo pachytene chromatin inactivation? Sci Rep 6:29949. https://doi.org/10.1038/srep29949
Merico V, Giménez MD, Vasco V, Zuccotti M, Searle JB, Hauffe HC, Garagna S, (2013) Chromosomal speciation in mice: a cytogenetic analysis of recombination. Chromosome Res 21:523-533. https://doi.org/10.1007/s10577-013-9377-5
Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, Heng HHQ, Spyropoulos B (1997) Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106:207–215
Mudry MD, Rahn IM, Solari AJ (2001) Meiosis and chromosome painting of sex chromosome systems in Ceboidea. Am J Primatol 54:65–78. https://doi.org/10.1002/ajp.1013
Parma P, Veyrunes F, Pailhoux E (2016) Sex reversal in non-human placental mammals. Sex Dev 10:326–344. https://doi.org/10.1159/000448361
Peters AH, Plug AW, van Vugt MJ, de Boer P (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosom Res 5:66–68
Rahn MI, Mudry M, Merani MS, Solari AJ (1996) Meiotic behavior of the X1X2Y1Y2 quadrivalent of the primate Alouatta caraya. Chromosom Res 4:350–356
Rahn MI, Noronha RC, Nagamachi CY, Pieczarka JC, Solari AJ, Sciurano RB (2016) Protein markers of synaptic behavior and chromatin remodeling of the neo-XY body in phyllostomid bats. Chromosoma 125:701–708. https://doi.org/10.1007/s00412-015-0566-1
Ratomponirina C, Viegas-Péquignot E, Dutrillaux B, Petter F, Rumpler Y (1986) Synaptonemal complexes in Gerbillidae: probable role of intercalated heterochromatin in gonosome-autosome translocations. Cytogenet Genome Res 43:161–167. https://doi.org/10.1159/000132315
Rens W, Grutzner F, O’Brien PCM et al (2004) From the cover: resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proc Natl Acad Sci 101:16257–16261. https://doi.org/10.1073/pnas.0405702101
Rens W, O’Brien PC, Grutzner F et al (2007) The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol 8:R243. https://doi.org/10.1186/gb-2007-8-11-r243
Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JMA (2010) Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol 20:2117–2123. https://doi.org/10.1016/j.cub.2010.11.010
Royo H, Prosser H, Ruzankina Y, Mahadevaiah SK, Cloutier JM, Baumann M, Fukuda T, Hoog C, Toth A, de Rooij DG, Bradley A, Brown EJ, Turner JMA (2013) ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes Dev 27:1484–1494. https://doi.org/10.1101/gad.219477.113
Saunders PA, Perez J, Rahmoun M, Ronce O, Crochet PA, Veyrunes F (2014) XY females do better than the XX in the African pygmy mouse, Mus minutoides. Evolution 68:2119–2127. https://doi.org/10.1111/evo.12387
Speed RM (1986) Oocyte development in XO foetuses of man and mouse: the possible role of heterologous X-chromosome pairing in germ cell survival. Chromosoma 94:115–124
Steinberg ER, Fortes VB, Rossi LF, Murer L, Lovato M, Merani MS, Mudry MD (2017) Cytogenetic characterization of brown howler monkeys, Alouatta guariba clamitans (Atelidae, Platyrrhini): meiotic confirmation of an X1X1X2X2X3X3/X1X2X3Y1Y2 sex chromosome system. Cytogenet Genome Res 151:131–140. https://doi.org/10.1159/000464375
Subramanian VV, Hochwagen A (2014) The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol 6:a016675. https://doi.org/10.1101/cshperspect.a016675
Taketo T, Naumova AK (2013) Oocyte heterogeneity with respect to the meiotic silencing of unsynapsed X chromosomes in the XY female mouse. Chromosoma 122:337–349. https://doi.org/10.1007/s00412-013-0415-z
Tarsounas M, Morita T, Pearlman RE, Moens PB (1999) RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol 147:207–220
Turner JM, Aprelikova O, Xu X et al (2004) BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol 14:2135–2142
Turner JM, Mahadevaiah SK, Fernandez-Capetillo O et al (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37:41–47
Vernet N, Szot M, Mahadevaiah SK, Ellis PJI, Decarpentrie F, Ojarikre OA, Rattigan A, Taketo T, Burgoyne PS (2014) The expression of Y-linked Zfy2 in XY mouse oocytes leads to frequent meiosis 2 defects, a high incidence of subsequent early cleavage stage arrest and infertility. Development 141:855–866. https://doi.org/10.1242/dev.091165
Veyrunes F, Perez J (2018) X inactivation in a mammal species with three sex chromosomes. Chromosoma 127:261–267. https://doi.org/10.1007/s00412-017-0657-2
Veyrunes F, Catalan J, Sicard B, Robinson TJ, Duplantier JM, Granjon L, Dobigny G, Britton-Davidian J (2004) Autosome and sex chromosome diversity among the African pygmy mice, subgenus Nannomys (Murinae; Mus). Chromosom Res 12:369–382. https://doi.org/10.1023/B:CHRO.0000034098.09885.e6
Veyrunes F, Watson J, Robinson TJ, Britton-Davidian J (2007) Accumulation of rare sex chromosome rearrangements in the African pygmy mouse, Mus (Nannomys) minutoides: a whole-arm reciprocal translocation (WART) involving an X-autosome fusion. Chromosom Res 15:223–230. https://doi.org/10.1007/s10577-006-1116-8
Veyrunes F, Catalan J, Tatard C, Cellier-Holzem E, Watson J, Chevret P, Robinson TJ, Britton-Davidian J (2010a) Mitochondrial and chromosomal insights into karyotypic evolution of the pygmy mouse, Mus minutoides, in South Africa. Chromosom Res 18:563–574. https://doi.org/10.1007/s10577-010-9144-9
Veyrunes F, Chevret P, Catalan J, Castiglia R, Watson J, Dobigny G, Robinson TJ, Britton-Davidian J (2010b) A novel sex determination system in a close relative of the house mouse. Proc Biol Sci 277:1049–1056. https://doi.org/10.1098/rspb.2009.1925
Veyrunes F, Perez J, Borremans B, Gryseels S, Richards LR, Duran A, Chevret P, Robinson TJ, Britton-Davidian J (2014) A new cytotype of the African pygmy mouse Mus minutoides in eastern Africa. Implications for the evolution of sex-autosome translocations. Chromosom Res 22:533–543. https://doi.org/10.1007/s10577-014-9440-x
Villemure M, Chen H-Y, Kurokawa M, Fissore RA, Taketo T (2007) The presence of X- and Y-chromosomes in oocytes leads to impairment in the progression of the second meiotic division. Dev Biol 301:1–13. https://doi.org/10.1016/j.ydbio.2006.10.034
Vozdova M, Ruiz-Herrera A, Fernandez J, Cernohorska H, Frohlich J, Sebestova H, Kubickova S, Rubes J (2016) Meiotic behaviour of evolutionary sex-autosome translocations in Bovidae. Chromosom Res 24:325–338. https://doi.org/10.1007/s10577-016-9524-x
Woods LM, Hodges CA, Baart E, Baker SM, Liskay M, Hunt PA (1999) Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J Cell Biol 145:1395–1406
Wu T, Lane SIR, Morgan SL, Jones KT (2018) Spindle tubulin and MTOC asymmetries may explain meiotic drive in oocytes. Nat Commun 9:2952. https://doi.org/10.1038/s41467-018-05338-7
Xu B, Obata Y, Cao F, Taketo T (2012) The presence of the Y-chromosome, not the absence of the second X-chromosome, alters the mRNA levels stored in the fully grown XY mouse oocyte. PLoS One 7:e40481. https://doi.org/10.1371/journal.pone.0040481
Acknowledgements
We thank Satoshi Namekawa for the anti-BRCA1 antibody, Marie Challe for her help in maintaining the breeding colony, and Julie Perez for genotyping some animals. We are especially indebted to the animal breeding facility of the University of Montpellier (CECEMA), the CytoEvol facilities of ISEM (labex CeMEB) and the imaging facility MRI, member of the national infrastructure France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04, «Investments for the future»).
Funding
This work was supported by grants from the Centre National pour la Recherche Scientifique (CNRS) and by the European Research Council (ERC) Executive Agency under the European Community’s Seventh Framework Programme (FP7/2007–2013 grant agreement no. 322788) to B.d.M. F.V. was funded by the French National Research Agency (ANR grant “SEXYMUS,” no. 10-JCJC-1605) and the Del Duca Foundation from Institut de France.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of a Special Issue on Recent advances in meiosis from DNA replication to chromosome segregation “edited by Valérie Borde and Francesca Cole, co-edited by Paula Cohen and Scott Keeney.”
Electronic supplementary material
Table S1
MLH1 foci in XX, XX* and X*Y pachytene oocytes in each individual mouse (DOCX 38 kb)
Figure S1
Synapsis of the sex-autosome quadrivalents in XX* and X*Y females. (a) Distribution of oocytes from XX, XX* and X*Y females at the indicated prophase I stages. The data from all mice of each genotype are pooled in Fig. 2a. Littermates are indicated by underlined or circled numbers (b) Distribution of pachytene oocytes from XX, XX* and X*Y females in the classes P1, P2, P3 and P4. P1, 1–2 fully asynapsed chromosome arms; P2, one partially asynapsed arm; P3, full synapsis with strong extended γH2AX signal on 1–2 arms; P4, full synapsis with no strong γH2AX signal. The females XX #1 and 2, XX* #2, 3 and 4, and X*Y #3 are littermates, as well as the females XX* #1 and X*Y #1. Data from all mice of each genotype are pooled in Fig. 2b. (c) Centromere signal distribution on the quadrivalent in fully synapsed (P3 and P4) pachytene oocytes from XX* and X*Y females. The data from all mice of each genotype are pooled in Fig. 3g. (PDF 143 kb)
Figure S2
BRCA1 immunostaining during meiotic prophase progression in XX females. (a-d) Chromosome spreads of oocytes from XX females stained with antibodies against centromere proteins (CEN, CREST serum, light blue), a meiotic chromosome axis marker (SYCP3, red) and BRCA1 (white or green). Stages are indicated on the left. Scale bar = 10 μm. (PDF 545 kb)
Figure S3
DMC1 immunostaining on meiotic chromosome spreads in one X*Y female. (a-d) Chromosome spreads of oocytes from one X*Y female (X*Y-1) were stained with antibodies against centromere proteins (CEN, CREST serum, blue), a meiotic chromosome axis marker (SYCP3, red) and DMC1 (green). P1, P3 and P4 classes as defined in the legend to Fig. 2. Enlarged view from each nucleus shows the sex chromosome-containing quadrivalent, with the corresponding drawing on the right panel. Recognizable chromosome arms are indicated by colored arrowheads, with the same color code as in Fig. 1. Scale bars = 10 μm. (e) Distribution of the nuclei with various patterns of DMC1 foci distribution among different classes of pachytene oocytes (P1, P2 and P3/P4). P3 and P4 nuclei were not distinguished due to the absence of γH2AX staining. P2 nuclei displaying DMC1 staining on both unsynapsed and nonhomologously synapsed regions were categorized according to the prevailing staining. (PDF 629 kb)
Figure S4
Numbers of MLH1 foci in XX, XX* and X*Y oocytes. (a) Number of MLH1 foci in pachytene nuclei displaying at least one MLH1 focus on each bivalent. Animals are the same as in Fig. S1, and the females XX #1 and 2, XX* #2, 3 and 4, and X*Y #3 are littermates. The data from all mice of each genotype are pooled in Fig. 5e. Oocyte numbers: XX-1, n = 47; XX-2, n = 58; XX*-2, n = 40; XX*-3, n = 63; XX*-4, n = 61; X*Y-2, n = 47; X*Y-3, n = 45. (PDF 150 kb)
Rights and permissions
About this article
Cite this article
Baudat, F., de Massy, B. & Veyrunes, F. Sex chromosome quadrivalents in oocytes of the African pygmy mouse Mus minutoides that harbors non-conventional sex chromosomes. Chromosoma 128, 397–411 (2019). https://doi.org/10.1007/s00412-019-00699-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-019-00699-4