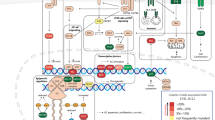
Abstract
Diffuse large B-cell lymphoma (DLBCL) is a common, genomically heterogenous disease that presents a clinical challenge despite the success of frontline regimens and second-line chimeric antigen receptor T-cell (CAR-T) therapy. Recently, genomic alterations and tumor microenvironment features associated with poor CAR-T response have been identified, namely those to the TP53 tumor suppressor gene. This retrospective analysis aimed to integrate various data to identify genomic partnerships capable of providing further clarity and actionable treatment targets within this population. Publicly available data were analyzed for differential expression based on TP53 and 24-month event-free survival (EFS24) status, revealing enrichments of the BRD4 bromodomain oncogene (p < 0.0001, p = 0.001). High-BRD4 and TP53 alterations were significantly associated with lower CDKN1A (p21) and TNFRSF10B (TRAIL-R2), a key tumor suppressor and CAR-T modulator, respectively. Significant loss of CD8 T-cell presence within low-TNFRSF0B (p = 0.0042) and altered-TP53 (p = 0.0424) patients showcased relevant outcome-associated tumor microenvironment features. Furthermore, reduced expression of CDKN1A was associated with low TNFRSF10B (FDR < 0.0001) and increased BRD4 interactant genes (FDR < 0.0001). Promisingly, in vitro MDM2 inhibition with Idasnutlin and TP53 reactivation via Eprenetapopt was able to renew TNFRSF10B protein expression. Additionally, applying the BRD4-degrading PROTAC ARV-825 and the CDK4/6 inhibitor Abemaciclib as single-agents and in synergistic combination significantly reduced TP53-altered DLBCL cell line viability. Our analysis presents key associations within a genomic network of actionable targets capable of providing clarity within the evolving precision CAR-T treatment landscape.





Similar content being viewed by others
Data availability
Data generated or analyzed during this study are included in this published article (and its supplemental information files) or are available publicly. Reasonable requests for additional data may be sent to the corresponding author.
References
Siegel RL, Miller KD, Wagle NS, Jemal A (2023) Cancer statistics, 2023. CA Cancer J Clin 73. https://doi.org/10.3322/caac.21763
Sehn LH, Salles G (2021) Diffuse Large B-Cell Lymphoma 384:842–858. https://doi.org/10.1056/NEJMra2027612.Diffuse
Fisher RI, Gaynor ER, Dahlberg S, Oken MM, Grogan TM, Mize EM et al (1993) Comparison of a standard regimen (CHOP) with three intensive chemotherapy regimens for advanced non-Hodgkin’s lymphoma. N Engl J Med 328:1002–1006. https://doi.org/10.1056/NEJM199304083281404
Tilly H, Morschhauser F, Sehn LH, Friedberg JW, Trněný M, Sharman JP et al (2022) Polatuzumab vedotin in previously untreated diffuse large B-cell lymphoma. N Engl J Med 386. https://doi.org/10.1056/nejmoa2115304
Crump M, Neelapu SS, Farooq U, Van Den Neste E, Kuruvilla J, Westin J et al (2017) Outcomes in refractory diffuse large B-cell lymphoma: results from the international SCHOLAR-1 study. Blood 130:1800–1808. https://doi.org/10.1182/blood-2017-03-769620
Ekstroem Smedby KE, Ekberg S, Eloranta S, Enblad G, Jerkeman M, Andersson P-O et al (2019) Treatment intensity, timing of relapse and outcome of 713 relapsed/refractory diffuse large B-cell lymphoma (DLBCL) in a population-based setting in Sweden. Blood 134. https://doi.org/10.1182/blood-2019-123785
Maurer MJ, Ghesquières H, Jais JP, Witzig TE, Haioun C, Thompson CA et al (2014) Event-free survival at 24 months is a robust end point for disease-related outcome in diffuse large B-cell lymphoma treated with immunochemotherapy. J Clin Oncol. https://doi.org/10.1200/JCO.2013.51.5866
June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC (2018) CAR T cell immunotherapy for human cancer. Science 80. https://doi.org/10.1126/science.aar6711
Buecklein V, Blumenberg V, Ackermann J, Schmidt C, Rejeski K, Mueller N et al (2020) Single-center experience with axicabtagene-ciloleucel (Axi-cel) and tisagenlecleucel (Tisa-cel) for relapsed/refractory diffuse large B-cell lymphoma: comparable response rates and manageable toxicity. Blood 136. https://doi.org/10.1182/blood-2020-142932
Sermer D, Batlevi C, Lia Palomba M, Shah G, Lin RJ, Perales MA et al (2020) Outcomes in patients with DLBCL treated with commercial CAR T cells compared with alternate therapies. Blood Adv 4. https://doi.org/10.1182/bloodadvances.2020002118
Locke FL, Neelapu SS, Bartlett NL, Siddiqi T, Chavez JC, Hosing CM, Ghobadi A, Budde LE, Bot A, Rossi JM, Jiang Y, Xue AX, Elias M, Aycock J, Wiezorek J, Go WY (2017) Phase 1 Results of ZUMA-1: A Multicenter Study of KTE-C19 Anti-CD19 CAR T Cell Therapy in Refractory Aggressive Lymphoma. Mol Ther 25(1):285–295. https://doi.org/10.1016/j.ymthe.2016.10.020
Brudno JN, Kochenderfer JN (2018) Chimeric antigen receptor T-cell therapies for lymphoma. Nat Rev Clin Oncol 15. https://doi.org/10.1038/nrclinonc.2017.128
Chong EA, Ruella M, Schuster SJ (2021) Five-year outcomes for refractory B-cell lymphomas with CAR T-cell therapy. N Engl J Med 384. https://doi.org/10.1056/nejmc2030164
Faramand R, Kotani H, Morrissey D, Yu B, Locke FL, Jain M et al (2019) Prediction of CAR T-related toxicities in R/R DLBCL patients treated with axicabtagene ciloleucel using point of care cytokine measurements. Biol Blood Marrow Transplant 25. https://doi.org/10.1016/j.bbmt.2018.12.827
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A et al (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503–511. https://doi.org/10.1038/35000501
Scott DW, Wright GW, Williams PM, Lih CJ, Walsh W, Jaffe ES et al (2014) Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. https://doi.org/10.1182/blood-2013-11-536433
Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A et al (2017) Genetic and functional drivers of diffuse large B cell lymphoma. Cell. https://doi.org/10.1016/j.cell.2017.09.027
Chapuy B, Stewart C, Dunford A, Kim J, Kamburov A, Redd R et al (2018) Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. https://doi.org/10.1038/s41591-018-0016-8
Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ et al (2018) Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. https://doi.org/10.1056/NEJMoa1801445
Lacy SE, Barrans SL, Beer PA, Painter D, Smith AG, Roman E et al (2020) Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood. https://doi.org/10.1182/blood.2019003535
Rushton C, Arthur SE, Alcaide M, Cheung M, Thomas N, Hilton LK et al (2019) Recurrent patterns of clonal evolution in relapsed-refractory DLBCL following treatment with R-CHOP. Blood 134:921–921. https://doi.org/10.1182/blood-2019-127399
Rushton CK, Arthur SE, Alcaide M, Cheung M, Jiang A, Coyle KM et al (2020) Genetic and evolutionary patterns of treatment resistance in relapsed B-cell lymphoma. Blood Adv 4. https://doi.org/10.1182/bloodadvances.2020001696
Rushton C, Alcaide M, Cheung M, Michaud NR, Daigle S, Rys RN et al (2020) The copy number landscape of relapsed and refractory diffuse large B-cell lymphoma. Blood 136. https://doi.org/10.1182/blood-2020-142871
Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L et al (2016) Genetic landscapes of relapsed and refractory diffuse large B-cell lymphomas. Clin Cancer Res 22:2290–2300. https://doi.org/10.1158/1078-0432.CCR-15-2123
Hilton LK, Ngu HS, Collinge B, Dreval K, Ben-Neriah S, Rushton CK et al (2022) Relapse timing is associated with distinct evolutionary dynamics and response to salvage therapy in DLBCL. Blood 140. https://doi.org/10.1182/blood-2022-160187
Kotlov N, Bagaev A, Revuelta MV, Phillip JM, Cacciapuoti MT, Antysheva Z et al (2021) Clinical and biological subtypes of b-cell lymphoma revealed by microenvironmental signatures. Cancer Discov 11. https://doi.org/10.1158/2159-8290.CD-20-0839
Steen CB, Luca BA, Esfahani MS, Azizi A, Sworder BJ, Nabet BY et al (2021) The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell 39. https://doi.org/10.1016/j.ccell.2021.08.011
Zhou Z, Sehn LH, Rademaker AW, Gordon LI, LaCasce AS, Crosby-Thompson A et al (2014) An enhanced International Prognostic Index (NCCN-IPI) for patients with diffuse large B-cell lymphoma treated in the rituximab era. Blood 123:837–842. https://doi.org/10.1182/blood-2013-09-524108
Shouse G, Kaempf A, Gordon MJ, Artz AS, Yashar D, Sigmund AM et al (2023) A validated composite comorbidity index predicts outcomes of CAR T-cell therapy in patients with diffuse large B cell lymphoma. Blood Adv. https://doi.org/10.1182/bloodadvances.2022009309
Jain MD, Ziccheddu B, Coughlin CA, Faramand R, Griswold AJ, Reid KM et al (2021) Genomic drivers of large B-cell lymphoma resistance to CD19 CAR-T therapy. Blood 138. https://doi.org/10.1182/blood-2021-148605
Shouval R, Alarcon Tomas A, Fein JA, Flynn JR, Markovits E, Mayer S et al (2021) Impact of TP53 genomic alterations in large B-cell lymphoma treated with CD19-chimeric antigen receptor T-cell therapy. J Clin Oncol. https://doi.org/10.1200/jco.21.02143
Sworder B, Kurtz DM, Macaulay C, Frank MJ, Alig S, Garofalo A et al (2019) Circulating DNA for molecular response prediction, characterization of resistance mechanisms and quantification of CAR T-cells during axicabtagene ciloleucel therapy. Blood 134. https://doi.org/10.1182/blood-2019-129015
Farinha P, Ennishi D, Mottok A, Ben-Neriah S, Meissner B, Boyle M et al (2019) TP53 expression correlates with TP53 mutations and is an independent predictor of clinical outcome in patients with DLBCL treated with R-CHOP. Blood 134. https://doi.org/10.1182/blood-2019-121943
Porpaczy E, Wohlfarth P, Königsbrügge O, Rabitsch W, Skrabs C, Staber P et al (2021) Influence of TP53 mutation on survival of diffuse large B-cell lymphoma in the CAR T-cell era. Cancers (Basel) 13. https://doi.org/10.3390/cancers13225592
Jardin F, Coiffier B (2013) TP53 and outcome in DLBCL: not only the coding region. Blood 121. https://doi.org/10.1182/blood-2013-04-493486
Monti S, Chapuy B, Takeyama K, Rodig SJ, Hao Y, Yeda KT et al (2012) Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell 22:359–372. https://doi.org/10.1016/j.ccr.2012.07.014
Lu TX, Young KH, Xu W, Li JY (2016) TP53 dysfunction in diffuse large B-cell lymphoma. Crit Rev Oncol Hematol 97. https://doi.org/10.1016/j.critrevonc.2015.08.006
Young KH, Weisenburger DD, Dave BJ, Smith L, Sanger W, Iqbal J et al (2007) Mutations in the DNA-binding codons of TP53, which are associated with decreased expression of TRAIL receptor-2, predict for poor survival in diffuse large B-cell lymphoma. Blood 110. https://doi.org/10.1182/blood-2007-02-072082
Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM et al (2012) Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood 120. https://doi.org/10.1182/blood-2012-05-433334
Lund S, Valentine Ngisa KW, Rutz A, Jinda Guidinger KTH (2022) Enrichment of TP53 alterations within GCB-like DNA subclassifications of diffuse large B-cell lymphoma after transition from de-novo to relapsed or refractory disease TO. Blood Res 17:190–191. https://doi.org/10.5045/br.2022.2022052
Dufva O, Koski J, Maliniemi P, Ianevski A, Klievink J, Leitner J et al (2020) Integrated drug profiling and CRISPR screening identify essential pathways for CAR T-cell cytotoxicity. Blood 135. https://doi.org/10.1182/blood.2019002121
Gu JJ, Thompson SJ, Mavis C, Barth MJ, Torka P, Hernandez-Ilizaliturri FJ (2019) Targeting MDM2 and XIAP by idasanutlin in diffuse large B-cell lymphoma. Blood 134. https://doi.org/10.1182/blood-2019-129009
Cluzeau T, Sebert M, Rahmé R, Cuzzubbo S, Lehmann-Che J, Madelaine I et al (2021) Eprenetapopt plus azacitidine in TP53-mutated myelodysplastic syndromes and acute myeloid Leukemia: a phase II study by the groupe francophone des Myélodysplasies (GFM). J Clin Oncol 39. https://doi.org/10.1200/JCO.20.02342
Sallman DA, DeZern AE, Garcia-Manero G, Steensma DP, Roboz GJ, Sekeres MA et al (2021) Eprenetapopt (APR-246) and azacitidine in TP53-mutant myelodysplastic syndromes. J Clin Oncol 39. https://doi.org/10.1200/JCO.20.02341.
Mondello P, Tadros S, Teater M, Fontan L, Chang AY, Jain N et al (2020) Selective inhibition of HDAC3 targets synthetic vulnerabilities and activates immune surveillance in lymphoma. Cancer Discov. https://doi.org/10.1158/2159-8290.CD-19-0116
Jain N, Hartert K, Tadros S, Fiskus W, Havranek O, Ma MCJ et al (2019) Targetable genetic alterations of TCF4 (E2-2) drive immunoglobulin expression in diffuse large B cell lymphoma. Sci Transl Med 11. https://doi.org/10.1126/scitranslmed.aav5599
Lu J, Qian Y, Raina K, Altieri M, Dong H, Wang J et al (2015) BRD4 degradation by protacs represents a more effective therapeutic strategy than BRD4 inhibitors in DLBCL. Blood 126. https://doi.org/10.1182/blood.v126.23.2050.2050
Yang H, Green MR (2020) Harnessing lymphoma epigenetics to improve therapies. Blood 136. https://doi.org/10.1182/BLOOD.2020006908
McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS et al (2012) EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492. https://doi.org/10.1038/nature11606
Tanaka Y, Momose S, Tabayashi T, Sawada K, Yamashita T, Higashi M et al (2020) Abemaciclib, a CDK4/6 inhibitor, exerts preclinical activity against aggressive germinal center-derived B-cell lymphomas. Cancer Sci 111. https://doi.org/10.1111/cas.14286
Xu-Monette ZY, Zhang H, Zhu F, Tzankov A, Bhagat G, Visco C et al (2020) A refined cell-of-origin classifier with targeted NGS and artificial intelligence shows robust predictive value in DLBCL. Blood Adv 4. https://doi.org/10.1182/bloodadvances.2020001949
Latif AL, Newcombe A, Li S, Gilroy K, Robertson NA, Lei X et al (2021) BRD4-mediated repression of p53 is a target for combination therapy in AML. Nat Commun 12. https://doi.org/10.1038/s41467-020-20378-8
Ouyang Z, Hardee J, Kundaje A, Zhang Y, Snyder M, Lacroute P (2013) STAT3 targets suggest mechanisms of aggressive tumorigenesis in diffuse large B-cell lymphoma. G3: Genes, Genomes, Genetics. https://doi.org/10.1534/g3.113.007674
Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP (2006) GenePattern 2.0 [2]. Nat Genet 38:500–501. https://doi.org/10.1038/ng0506-500
Gould J, Getz G, Monti S, Reich M, Mesirov JP (2006) Comparative gene marker selection suite. Bioinformatics. https://doi.org/10.1093/bioinformatics/btl196
Chen J, Bardes EE, Aronow BJ, Jegga AG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. https://doi.org/10.1093/nar/gkp427
Vallania F, Tam A, Lofgren S, Schaffert S, Azad TD, Bongen E et al (2018) Leveraging heterogeneity across multiple datasets increases cell-mixture deconvolution accuracy and reduces biological and technical biases. Nat Commun 9. https://doi.org/10.1038/s41467-018-07242-6
Ianevski A, Giri AK, Aittokallio T (2021) SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res 48. https://doi.org/10.1093/NAR/GKAA216
Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MGM, Qi J et al (2013) Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24:777–790. https://doi.org/10.1016/j.ccr.2013.11.003
Ceribelli M, Hou ZE, Kelly PN, Huang DW, Wright G, Ganapathi K et al (2016) A druggable TCF4- and BRD4-dependent transcriptional network sustains malignancy in blastic plasmacytoid dendritic cell neoplasm. Cancer Cell 30:764–778. https://doi.org/10.1016/j.ccell.2016.10.002
Kreis NN, Louwen F, Yuan J (2019) The multifaceted p21 (Cip1/Waf1/CDKN1A) in cell differentiation, migration and cancer therapy. Cancers (Basel) 11. https://doi.org/10.3390/cancers11091220
Mensah AA, Kwee I, Gaudio E, Rinaldi A, Ponzoni M, Cascione L et al (2015) Novel HDAC inhibitors exhibit pre-clinical efficacy in lymphoma models and point to the importance of CDKN1A expression levels in mediating their anti-tumor response. Oncotarget 6. https://doi.org/10.18632/oncotarget.3239
Fiskus W, Cai T, DiNardo CD, Kornblau SM, Borthakur G, Kadia TM et al (2019) Superior efficacy of cotreatment with BET protein inhibitor and BCL2 or MCL1 inhibitor against AML blast progenitor cells. Blood Cancer J 9. https://doi.org/10.1038/s41408-018-0165-5
Andrieu G, Belkina AC, Denis GV (2016) Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today Technol 19:45–50. https://doi.org/10.1016/j.ddtec.2016.06.004
Wu S, Jiang Y, Hong Y, Chu X, Zhang Z, Tao Y et al (2021) BRD4 PROTAC degrader ARV-825 inhibits T-cell acute lymphoblastic leukemia by targeting “Undruggable” Myc-pathway genes. Cancer Cell Int 21. https://doi.org/10.1186/s12935-021-01908-w
Acknowledgements
The views expressed in the submitted article are those of the authors and not an official position of the institution or funder. Dr. Javeed Iqbal and Dr. Alyssa Bouska from the University of Nebraska Medical Center kindly gifted the DHL16, Karpas-422, Ly3, and U2932 cell lines. Dr. Anne Novak from the Mayo Clinic, Rochester kindly gifted the DHL6 cell line.
Funding
This study was supported by the MNSU Faculty Research Grant (Minnesota State Colleges and Universities system), MNSU Undergraduate Research Center Foundation grants, and the Minnesota State University Mankato faculty startup fund. The funding sources were not involved in the study design, collection, analysis, interpretation of data, writing of the report, or decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
Author contributions are as follows: A.F., J. U., K.W., A.R., S.L., J.G., A.P., J. H., A.H., and K.T.H. performed investigations and analyses. K.T.H. and A.F. were responsible for revision. K.T.H. was responsible for conceptualization, data curation, formal analysis, funding acquisition, writing, and supervision.
Corresponding author
Ethics declarations
Ethical approval
This is an observational study. All de-identified datasets were assembled from previously published and publicly available analyses.
Competing interests
The authors declare no competing interests.
Copyright transfer agreement
Each author has contributed to this manuscript substantially and intellectually and shares the public responsibility for its contents. Each author warrants that his/her contributions to this manuscript are original works not published wholly or partly elsewhere, except in the form of an abstract; that he/she will not submit it to other journals except in the case of editorial rejection and in the case of duplicate publication that was approved by both editors-in-chief of the first and second journals; and that the manuscript contains nothing unlawful, invading the right of privacy, or infringing a proprietary right, so that the journal should not be responsible for such legal affairs. Each author warrants the transfer of the copyright, interest, authorship, and all rights regarding this manuscript to the publisher, in case of publication. The copyright agreement has been agreed upon by all parties at the time of May 2023.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Aidan L. Forberg, Jordan Unrau and Kennedee S. Weber are co-authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Forberg, A.L., Unrau, J., Weber, K.S. et al. Integrative analyses reveal outcome-associated and targetable molecular partnerships between TP53, BRD4, TNFRSF10B, and CDKN1A in diffuse large B-cell lymphoma. Ann Hematol 103, 199–209 (2024). https://doi.org/10.1007/s00277-023-05478-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-023-05478-x