Abstract
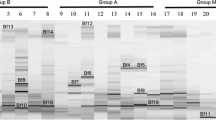
The beneficial effects of Bifidobacteria on health have been widely accepted. Patients with chronic liver disease have varying degrees of intestinal microflora imbalance with a decrease of total Bifidobacterial counts. Since different properties have been attributed to different Bifidobacterium species and there is no information available for the detailed changes in the genus Bifidobacterium in patients with chronic liver disease heretofore, it is meaningful to investigate the structure of this bacterium at the species level in these patients. The aim of this study was to characterize the composition of intestinal Bifidobacterium in patients with hepatitis B virus-induced chronic liver disease. Nested-PCR-based denaturing gradient gel electrophoresis (PCR-DGGE), clone library, and real-time quantitative PCR were performed on the fecal samples of 16 patients with chronic hepatitis B (CHB patients), 16 patients with hepatitis B virus-related cirrhosis (HBV cirrhotics), and 15 healthy subjects (Controls). Though there was no significant difference in the diversity among the three groups (P = 0.196), Bifidobacterium dentium seems to be specifically enhanced in patients as the PCR-DGGE profiles showed, which was further validated by clone library and real-time quantitative PCR. In contrast to the B. dentium, Bifidobacterium catenulatum/Bifidobacterium pseudocatenulatum were detected less frequently in the predominant profile and by quantitative PCR in HBV cirrhotics than in the controls, and the level of this species was also significantly different between these two groups (P = 0.023). Although having no quantitative difference among the three groups, Bifidobacterium longum was less commonly detected in HBV cirrhotics than in CHB patients and Controls by quantitative PCR (P = 0.011). Thus, the composition of intestinal Bifidobacterium was deeply altered in CHB and HBV cirrhotic patients with a shift from beneficial species to opportunistic pathogens. The results provide further insights into the dysbiosis of the intestinal microbiota in patients with hepatitis B virus-induced chronic liver disease and might potentially serve as guidance for the probiotics interventions of these diseases.



Similar content being viewed by others
References
Lavanchy D (2004) Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat 11:97–107
Garcia-Tsao G, Wiest R (2004) Gut microflora in the pathogenesis of the complications of cirrhosis. Best Pract Res Clin Gastroenterol 18:353–372
Norman K, Pirlich M (2008) Gastrointestinal tract in liver disease: which organ is sick? Curr Opin Clin Nutr Metab Care 11:613–619
Riordan SM, Williams R (2006) The intestinal flora and bacterial infection in cirrhosis. J Hepatol 45:744–757
Trebichavsky I, Rada V, Splichalova A, Splichal I (2009) Cross-talk of human gut with bifidobacteria. Nutr Rev 67:77–82
Mitsuoka T (1990) Bifidobacteria and their role in human health. J Ind Microbiol 6:263–267
Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, Jedynak-Wasowicz U, Sladek M, Pieczarkowski S, Adamski P, Kochan P, Heczko PB (2009) Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J Gastroenterol 15:5287–5294
Kerckhoffs AP, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K, Akkermans LM (2009) Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol 15:2887–2892
Li L, Wu Z, Ma W, Yu Y, Chen Y (2001) Changes in intestinal microflora in patients with chronic severe hepatitis. Chin Med J (Engl) 114:869–872
Zhao HY, Wang HJ, Lu Z, Xu SZ (2004) Intestinal microflora in patients with liver cirrhosis. Chin J Dig Dis 5:64–67
Lu H, Wu Z, Xu W, Yang J, Chen Y, Li L (2011) Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection: intestinal microbiota of HBV cirrhotic patients. Microb Ecol 61(3):693–703
Lopez P, Gueimonde M, Margolles A, Suarez A (2010) Distinct Bifidobacterium strains drive different immune responses in vitro. Int J Food Microbiol 138:157–165
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature 457:480–484
Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, van Sinderen D, Ventura M (2009) Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J 3:745–751
Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y (2008) Imbalances in faecal and duodenal Bifidobacterium species composition in active and non-active coeliac disease. BMC Microbiol 8:232
Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P (2009) Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101:541–550
Martin R, Jimenez E, Heilig H, Fernandez L, Marin ML, Zoetendal EG, Rodriguez JM (2009) Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol 75:965–969
Kuhbacher T, Ott SJ, Helwig U, Mimura T, Rizzello F, Kleessen B, Gionchetti P, Blaut M, Campieri M, Folsch UR, Kamm MA, Schreiber S (2006) Bacterial and fungal microbiota in relation to probiotic therapy (VSL#3) in pouchitis. Gut 55:833–841
Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R (2004) Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol 70:167–173
Zhuang H (2007) Guideline on prevention and treatment of chronic hepatitis B in China (2005). Chinese Med J-Peking 120:2159–2173
Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R (1973) Transection of the oesophagus for bleeding oesophageal varices. Br J Surg 60:646–649
Nielsen DS, Moller PL, Rosenfeldt V, Paerregaard A, Michaelsen KF, Jakobsen M (2003) Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon. Appl Environ Microbiol 69:7545–7548
Satokari RM, Vaughan EE, Akkermans AD, Saarela M, de Vos WM (2001) Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol 67:504–513
Simpson JM, McCracken VJ, Gaskins HR, Mackie RI (2000) Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl Environ Microbiol 66:4705–4714
Ritchie LE, Steiner JM, Suchodolski JS (2008) Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol Ecol 66:590–598
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Pei CX, Liu Q, Dong CS, Li H, Jiang JB, Gao WJ (2010) Diversity and abundance of the bacterial 16S rRNA gene sequences in forestomach of alpacas (Lama pacos) and sheep (Ovis aries). Anaerobe 16:426–432
Sanz Y, Sanchez E, Marzotto M, Calabuig M, Torriani S, Dellaglio F (2007) Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol Med Microbiol 51:562–568
Sheth AA, Garcia-Tsao G (2008) Probiotics and liver disease. J Clin Gastroenterol 42(Suppl 2):S80–S84
Malaguarnera M, Greco F, Barone G, Gargante MP, Toscano MA (2007) Bifidobacterium longum with fructo-oligosaccharide (FOS) treatment in minimal hepatic encephalopathy: a randomized, double-blind, placebo-controlled study. Dig Dis Sci 52:3259–3265
Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H (1999) Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol 65:4506–4512
Beighton D, Gilbert SC, Clark D, Mantzourani M, Al-Haboubi M, Ali F, Ransome E, Hodson N, Fenlon M, Zoitopoulos L, Gallagher J (2008) Isolation and identification of bifidobacteriaceae from human saliva. Appl Environ Microbiol 74:6457–6460
Hojo K, Nagaoka S, Murata S, Taketomo N, Ohshima T, Maeda N (2007) Reduction of vitamin K concentration by salivary Bifidobacterium strains and their possible nutritional competition with Porphyromonas gingivalis. J Appl Microbiol 103:1969–1974
Mantzourani M, Gilbert SC, Sulong HN, Sheehy EC, Tank S, Fenlon M, Beighton D (2009) The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res 43:308–313
Nakajo K, Takahashi N, Beighton D (2010) Resistance to acidic environments of caries-associated bacteria: Bifidobacterium dentium and Bifidobacterium longum. Caries Res 44:431–437
Gueimonde M, Debor L, Tolkko S, Jokisalo E, Salminen S (2007) Quantitative assessment of faecal bifidobacterial populations by real-time PCR using lanthanide probes. J Appl Microbiol 102:1116–1122
Ventura M, Turroni F, Zomer A, Foroni E, Giubellini V, Bottacini F, Canchaya C, Claesson MJ, He F, Mantzourani M, Mulas L, Ferrarini A, Gao B, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Gupta RS, Zhang Z, Beighton D, Fitzgerald GF, O’Toole PW, van Sinderen D (2009) The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet 5:e1000785
Fardini Y, Chung P, Dumm R, Joshi N, Han YW (2010) Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun 78:1789–1796
Li YT, Wang L, Chen Y, Chen YB, Wang HY, Wu ZW, Li LJ (2010) Effects of gut microflora on hepatic damage after acute liver injury in rats. J Trauma 68:76–83
Xing HC, Li LJ, Xu KJ, Shen T, Chen YB, Sheng JF, Chen Y, Fu SZ, Chen CL, Wang JG, Yan D, Dai FW, Zheng SS (2006) Protective role of supplement with foreign Bifidobacterium and Lactobacillus in experimental hepatic ischemia-reperfusion injury. J Gastroenterol Hepatol 21:647–656
Romond MB, Haddou Z, Mielcareck C, Romond C (1997) Bifidobacteria and human health: regulatory effect of indigenous bifidobacteria on Escherichia coli intestinal colonization. Anaerobe 3:131–136
Matsumoto T, Ishikawa H, Tateda K, Yaeshima T, Ishibashi N, Yamaguchi K (2008) Oral administration of Bifidobacterium longum prevents gut-derived Pseudomonas aeruginosa sepsis in mice. J Appl Microbiol 104:672–680
Han SY, Huh CS, Ahn YT, Lim KS, Baek YJ, Kim DH (2005) Hepatoprotective effect of lactic acid bacteria, inhibitors of beta-glucuronidase production against intestinal microflora. Arch Pharm Res 28:325–329
Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL (2010) Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341
Kulagina EV, Shkoporov AN, Kafarskaia LI, Khokhlova EV, Volodin NN, Donskikh EE, Korshunova OV, Efimov BA (2010) Molecular genetic study of species and strain variability in bifidobacteria population in intestinal microflora of breast-fed infants and their mothers. Bull Exp Biol Med 150:61–64
Delgado S, Suarez A, Mayo B (2006) Bifidobacterial diversity determined by culturing and by 16S rDNA sequence analysis in feces and mucosa from ten healthy Spanish adults. Dig Dis Sci 51:1878–1885
Acknowledgments
This study was supported by the National Basic Research Program (973) of China (2007CB513003 and 2009CB522406) and Zhejiang Public Health Bureau Fund (2008QN010). We thank Dr. Wei Chen for critical reading and useful suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, M., Wang, B., Fu, Y. et al. Changes of Fecal Bifidobacterium Species in Adult Patients with Hepatitis B Virus-Induced Chronic Liver Disease. Microb Ecol 63, 304–313 (2012). https://doi.org/10.1007/s00248-011-9925-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9925-5