Abstract
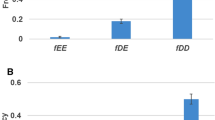
In the yeast or nematode, the proportion of essential genes in duplicates is lower than in singletons (single-copy genes), due to the functional redundancy. One may expect that it should be the same in the mouse genome. However, based on the publicly available mouse knockout data, it was observed that the proportion of essential genes in duplicates is similar to that in singletons. The most straightforward interpretation, as claimed in a recent study, is that duplicate genes may have a negligible role in the mouse genetic robustness. Here we show that in the current mouse knockout dataset, recently duplicated genes have been highly underrepresented, leading to an overestimation of the proportion of essential genes in duplicates. After estimating the duplication time of mouse duplication events, we have developed a simple bias-correcting procedure and shown that the bias-corrected proportion of essential genes in mouse duplicates is significantly lower than that in singletons.


Similar content being viewed by others
References
Conant GC, Wagner A (2004) Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc Biol Sci 271:89–96
Dean EJ, Davis JC, Davis RW, Petrov DA (2008) Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet 4(7):e1000113
Gu X (2003) Evolution of duplicate genes versus genetic robustness against null mutations. Trends Genet 19:354–356
Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H (2002) Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol 19:256–262
Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W-H (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421:63–66
Harrison R, Papp B, Pal C, Oliver SG, Delneri D (2007) Plasticity of genetic interactions in metabolic networks of yeast. Proc Natl Acad Sci USA 104:2307–2312
Ihmels J, Collins SR, Schuldiner M, Krogan N, Weissman JS (2007) Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Syst Biol 3:86
Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231–237
Liang H, Li W-H (2007) Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet 23:375–378
Liao B-Y, Zhang J (2007) Mouse duplicate genes are as essential as singletons. Trends Genet 23:378–381
Pearson WR (2000) Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol 132:185–219
Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906
Acknowledgments
This work was supported in part by grants (30428003 to X.G., 30700140 to Z.S.) from the National Natural Science Foundation of China, and grants from Iowa State University and Fudan University (to X.G.), and supported by the Shanghai Leading Academic Discipline Project (Project No. B111). We are grateful to Wen-Hsiung Li, Han Liang, and Jianzhi Zhang for critical comments on the early version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Su, Z., Gu, X. Predicting the Proportion of Essential Genes in Mouse Duplicates Based on Biased Mouse Knockout Genes. J Mol Evol 67, 705–709 (2008). https://doi.org/10.1007/s00239-008-9170-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9170-9