Abstract
Aims/hypothesis
Increasing evidence suggests that environmental factors changing the normal colonisation pattern in the gut strongly influence the risk of developing autoimmune diabetes. The aim of this study was to investigate, both during infancy and adulthood, whether treatment with vancomycin, a glycopeptide antibiotic specifically directed against Gram-positive bacteria, could influence immune homeostasis and the development of diabetic symptoms in the NOD mouse model for diabetes.
Methods
Accordingly, one group of mice received vancomycin from birth until weaning (day 28), while another group received vancomycin from 8 weeks of age until onset of diabetes. Pyrosequencing of the gut microbiota and flow cytometry of intestinal immune cells was used to investigate the effect of vancomycin treatment.
Results
At the end of the study, the cumulative diabetes incidence was found to be significantly lower for the neonatally treated group compared with the untreated group, whereas the insulitis score and blood glucose levels were significantly lower for the mice treated as adults compared with the other groups. Mucosal inflammation was investigated by intracellular cytokine staining of the small intestinal lymphocytes, which displayed an increase in cluster of differentiation (CD)4+ T cells producing pro-inflammatory cytokines in the neonatally treated mice. Furthermore, bacteriological examination of the gut microbiota composition by pyrosequencing revealed that vancomycin depleted many major genera of Gram-positive and Gram-negative microbes while, interestingly, one single species, Akkermansia muciniphila, became dominant.
Conclusions/interpretation
The early postnatal period is a critical time for microbial protection from type 1 diabetes and it is suggested that the mucolytic bacterium A. muciniphila plays a protective role in autoimmune diabetes development, particularly during infancy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
NOD mice spontaneously start to develop insulitis at 3 to 5 weeks of age [1] and, in addition to host genetics, intestinal-related environmental factors, in particular the gut microbiota, appear central to the development of type 1 diabetes [2].
The gut contains several hundred species of microorganism that make essential contributions to nutrient absorption, intestinal integrity and defence against colonisation by pathogenic bacteria [3]. In addition, recent publications have revealed the importance of specific intestinal bacteria in promoting regulatory immunity [4, 5], whereas other microbial species have more pro-inflammatory properties, such as the Gram-positive segmented filamentous bacteria (SFB) [6]. Dendritic cells (DCs) in the intestinal mucosa communicate with the microbiota through innate receptors such as toll-like receptors (TLRs) [7]. In a favourable microbial environment DCs develop tolerogenic properties and are capable of inducing different subsets of regulatory T (Treg) cells to keep the homeostatic balance between tolerance and immunity in check. Studies show conflicting results on the extent to which microbial epitopes stimulate regulatory immunity [8, 9], but failure to shape proper tolerogenic immune mechanisms as a result of dysbiosis has been suggested as a potential causative link to diabetes [10].
Supporting evidence of a protective role of intestinal bacteria against type 1 diabetes development has been found in the fact that type 1 diabetes incidence in diabetes prone rodents depends on their microbial status [11]. Germ-free NOD mice do not differ substantially from conventional NOD mice in the incidence of type 1 diabetes, but they have accentuated insulitis, increased levels of Il17 (also known as Il17a) mRNA in the colon, as well as increased numbers of T helper (Th) 17 and Th1 cells in their mesenteric lymph nodes (MLNs) [12]. Lack of the myeloid differentiation primary response gene (88) (MYD88) receptor, used by most TLRs, totally prevents diabetes development in NOD mice [13], and this has been linked to a decreased Firmicutes (Gram positive)/Bacteroidetes (Gram negative) ratio. Furthermore, previous studies with animal models for type 1 diabetes have shown an anti-diabetogenic effect of a gut microbiota composition with fewer Gram-positive microaerophilic bacteria [14, 15]. However, the impact of specific gut microbiota composition on the development of type 1 diabetes during different periods of life remains to be identified.
The infant gut displays a Th2 skewed cytokine profile that favours induction of immunological ignorance toward dietary and bacterial antigens [16–18]. Furthermore, previous findings suggest that immunomodulators of the neonatal gut immune system play a particular role in controlling diabetes development [19, 20]. According to the hygiene hypothesis, decreased bacterial exposure in early life leads to defective Treg cell induction, and is thus responsible for an autoimmune initiation of inflammatory cytokine production [2, 21, 22]. There are, thus, good reasons to believe that alterations in the early microbial colonisation of the gut by neonatal antibiotic treatment will have an immense impact on diabetes progression later in life. In particular, treatment with vancomycin, a glycopeptide antibiotic that targets Gram-positive bacteria by inhibiting their cell wall synthesis, has been shown to alter important mucosal inflammatory markers [23].
Methods
The experiments were carried out in accordance with the Council of Europe Convention European Treaty Series (ETS) 123 on the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes, and the Danish Animal Experimentation Act (LBK 1306 from 23/11/2007). The study was approved by the Animal Experiments Inspectorate, Ministry of Justice, Denmark.
Animals
Inbred NOD/BomTac mice (Taconic, Lille Skensved, Denmark) were purchased and mated, and their female offspring group-housed (five mice/cage) at our animal facility (Faculty of Life Science, University of Copenhagen, Frederiksberg, Denmark) under standard conditions. Mice had free access to Altomin 1324 diet (Brogaarden, Lynge, Denmark) and water.
Group size was between 15 and 21 mice per group. From 10 weeks of age, measurements of tail blood glucose commenced twice a week. A mouse was considered diabetic and killed when blood glucose levels exceeded 12 mmol/l on 2 consecutive days. At the age of 30 weeks, all remaining mice were killed by cervical dislocation.
Antibiotic treatment
In one group, mice were provided with vancomycin hydrochloride (0.5 g/l; ThermoFisher Scientific, Waltham, MA, USA) in the water from the age of 8 weeks and this was continued until diabetes onset (adult vancomycin treatment). Another group received vancomycin hydrochloride from birth until weaning (day 28) by oral feeding daily using a pipette (neonatal vancomycin treatment). Vancomycin hydrochloride was dissolved in water and given to the pups in a dose equivalent to the dose provided to an adult mouse (83 mg kg−1 day−1) based on calculation of their metabolic weight ratio and an expected daily water consumption of 5 ml per adult mouse (30 g). In addition, their mothers received vancomycin hydrochloride (0.5 g/l) added into the drinking water. After weaning, the neonatally treated mice received pure tap water (recovering). A control group only received pure tap water. Only the neonatally treated mice were orally fed with a suspension but all mice (untreated and treated) were handled daily. Bottles with water and vancomycin were changed twice weekly.
Histology
Haematoxylin and eosin stained pancreas sections were evaluated for insulitis score in a blinded fashion by two scientists. Lymphocytic infiltration was graded as follows: 0, no infiltration; 1, intact islets but with few mononuclear cells surrounding the islets; 2, peri-insulitis; 3, islet infiltration below 50%; 4, islet infiltration above 50%. Twenty-five islets were scored for each of six mouse models for diabetes in each group.
Cell isolation and flow cytometry
Immediately after the mice were killed, organs were placed in cold PBS. Single cell suspensions from spleen, MLNs and pancreatic lymph nodes (PLNs) were prepared by passing the freshly isolated organs through a 100 μm nylon mesh. After 5 min in erythrocyte lysing solution (0.15 mol/l NH4Cl, 10 mmol/l KHCO3, 1 mmol/l EDTA monosodium pH 7.3), cells were washed in cold PBS and resuspended in culture medium (RPMI 1640 supplemented with 10% FCS, 2 mmol/l l-glutamine and 2 mmol/l penicillin and streptomycin; all from Sigma-Aldrich, St Louis, MO, USA). Surface staining of DCs was performed for 30 min with the appropriate antibodies.
After removal of residual mesenteric fat from the entire small intestine, intestines were opened longitudinally, washed with cold PBS, and cut in 1 cm pieces. The pieces were incubated in 5 ml PBS containing 2 mmol/l EDTA for 20 min at 37°C with agitation (50 rpm). Subsequently, the fragments were shaken intensively to detach the epithelial cells and passed through a 70 μm cell strainer. The remaining fragments were minced using razor blades and placed in 10 ml fresh digestion solution (RPMI 1640 supplemented with 10% FCS, 100 U/ml collagenase and 50 U/ml DNase I; all from Sigma-Aldrich). After 45 min digestion at 37°C with rotation (200 rpm), the supernatant fraction was passed through a 100 μm cell strainer, washed in RPMI 1640 and resuspended in 8 ml of 40% Percoll and overlaid on 5 ml of 80% Percoll (GE Healthcare, Broendby, Denmark). Lymphocytes were collected at the interface of the gradient following centrifugation for 25 min at 600 g at room temperature and resuspended in culture medium. For Treg staining, cells were first surface stained, then fixed, permeabilised, and stained for intracellular forkhead box P3 (FOXP3) according to the manufacturer’s protocol (eBiosciences, San Diego, CA, USA). For intracellular cytokine staining, cells were incubated for 4 h with 50 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) and 750 ng/ml ionomycin (Sigma-Aldrich) in the presence of GolgiStop (0.7 μl/ml; BD Biosciences, San Jose, CA, USA) in an incubator at 37°C. Cells were stained and fixed using the same protocol as described for Tregs. All antibodies were purchased from eBiosciences. Analysis was performed using an Accuri C6 flow cytometer (Accuri Cytometers, Ann Arbor, MI, USA).
Bacterial analysis by denaturing gradient gel electrophoresis
The samples analysed include the faeces samples aseptically collected for each group when the mice were killed and, for the neonatal treatment group, also at the age of 4 weeks during antibiotic administration. The samples were stored at −80°C until further processing. A detailed description of the analysis by denaturing gradient gel electrophoresis (DGGE) is given elsewhere [24]. Briefly, cellular DNA was extracted using a QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and stored at −40°C until analysis. The DNA was amplified by means of PCR, using primers specific to the V3 region of the 16S ribosomal RNA (rRNA) gene. Thereafter, amplicons were separated by means of DGGE on polyacrylamide gel containing a 30–65% chemical gradient (100% corresponds to 7 mol/l urea and 40% formamide) enabling separation of the DNA amplicons based on sequence differences. After 16 h of electrophoresis, the gels were stained with SYBR gold and photographed.
Tag-encoded pyrosequencing
Amplicons (466 bp) flanking the V3 and V4 regions of the 16S rRNA gene were amplified using the primers detailed in electronic supplementary material (ESM) Table 1 [25] followed by a second round of PCR where primers with adapters and tags were used [26]. PCR amplification of the 16S rRNA gene and purification and pyrosequencing of amplified PCR products were carried out as previously described [27]. The amplified fragments with adapters and tags were quantified using a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) and mixed in approximately equal concentrations (14 × 107 copies per μl) to ensure equal representation of each sample. A two-region 454 sequencing run was performed on a GS FLX Titanium Pico TiterPlate (70 × 75) using a GS FLX Titanium Sequencing Kit XLR70 according to the manufacturer’s instructions (Roche Diagnostics, Indianapolis, IN, USA). Data from 454 tag-encoded sequencing were analysed using the Pyrosequencing Pipeline from the Ribosomal Database Project (RDP). Sequence quality control, sorting and trimming were performed for quality score >20 and minimum sequence length of 300 bp. Classification of sequences into taxa was carried out using RDP10 (RDP database, confidence threshold 80%). Data have been submitted to the NCBI Sequence Read Archive under study number SRA047328.1. Sequences from the individual samples were analysed as four groups. Two groups from samples collected during vancomycin treatment either as pups or adults, one group representing untreated mice and one group representing mice that had recovered from neonatal vancomycin treatment. After the annotation step was completed, numbers of sequences featured with a hit were normalised according to the size of each individual metagenome. The Shannon index and Chao1 estimator, as well as the Rarefaction tool, preceded by alignment and complete linkage clustering, were used to evaluate species richness and diversity among the samples. All indexes were computed for a cut-off distance of 3%, which is commonly used as the species determining distance [28].
Statistics
GraphPad Prism version 5.02 (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. Cumulative diabetes incidence was calculated using the Kaplan–Meier estimation while statistical significance was evaluated by the logrank test. Other differences were estimated by one-way ANOVA with Dunnett’s post test or the Kruskal–Wallis test on data that did not assume Gaussian distributions. A p value of less than 0.05 was considered significant. DGGE profile comparison was performed by the Dice similarity coefficient with a band position tolerance and optimisation of 1% using the unweighted pair group method with arithmetic averages clustering algorithm (UPGMA) and by principal component analysis (PCA) using BioNumerics Version 4.5 (Applied Maths, Sint-Martens-Latem, Belgium).
Results
Vancomycin suppresses clinical onset of diabetes in NOD mice
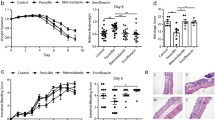
The percentage of diabetic mice at the end of the study was 93% for the untreated mice (n = 15), 73% for the mice treated with vancomycin from 8 weeks of age (n = 15) and 75% for the mice treated with vancomycin neonatally (n = 21). The mean diabetes onset time was 146 ± 8 (mean ± SE) days for the untreated mice and 161 ± 10 and 168 ± 7 days for the mice treated with vancomycin as adults and pups, respectively. Although vancomycin treatment seems to have affected diabetes development in both groups, the cumulative diabetes incidence was only significantly lower for the mice treated with vancomycin before weaning compared with the untreated mice (p < 0.05) (Fig. 1).
Cumulative diabetes incidence in NOD female mice. Mice received pure water (n = 15, solid line) or vancomycin treatment as adults VA from 8 weeks of age (n = 15, dashed line) or vancomycin treatment as pups VP from birth and until 4 weeks of age (n = 21, dotted line). Mice were diagnosed as diabetic and killed when blood glucose levels exceeded 12 mmol/l on two consecutive days. Comparisons of survival curves were tested by logrank test; p = 0.026, VP vs untreated; p = 0.169, VA vs untreated
Blood glucose values at the time the mice were killed were lower for the vancomycin-treated groups compared with the untreated group; however this was only significant for the adult treated group (p < 0.05) (Table 1). Average insulitis scores were similar in the untreated and the neonatally treated group but the insulitis score was significantly lower in the adult treated group compared with the other groups (p < 0.01) (Table 2). The non-diabetic insulitis scores for the adult treated mice were significantly lower than the average insulitis score in all groups of diabetic mouse models (p < 0.001).
Furthermore, no significant differences in weight gain were observed between the groups within the observational period.
Characterisation of the gut microbiota by DGGE
PCA of the DGGE profiles revealed clearly distinguishable main clusters separating the faeces samples from mice treated with vancomycin and untreated mice (Fig. 2). Furthermore, significant separation was also found between mice treated either as adults or pups.
PCA plots based on DGGE profiles of 16S rRNA gene PCR-derived amplicons of faeces samples confirmed the effectiveness of vancomycin treatment on the gut microbiota composition. (a) Untreated control mice (round balls) and mice during vancomycin treatment as pups (cylinders) or as adults (squares). ANOVA based on the first (X), second (Y) and third (Z) principal component (PC) revealed a significant difference in PC1 (explaining 43.3% of the variance) values between untreated and vancomycin-treated groups (p < 0.001) and significant differences in PC2 (explaining 10.4% of the variance) values among all of the groups (p < 0.001). (b) Untreated mice (round balls) and mice that have recovered from vancomycin treatment as pups (diamonds). ANOVA based on the first (X), second (Y) and third (Z) PC revealed a significant difference in PC1 (explaining 19.1% of the variance) values between untreated and recovered mice (p < 0.01)
PCA of the DGGE profiles from faeces samples collected from untreated mice and mice that had recovered from neonatal vancomycin treatment also revealed a significant separation of the two groups. These samples were collected when the mice were killed more than 12 weeks after neonatal vancomycin treatment, and this demonstrates how the gut microbiota composition did not completely recover after vancomycin treatment ended (Fig. 2).
Characterisation of the gut microbiota by pyrosequencing
The overall number of reads generated by 454 sequencing for 66 samples scored 913,455. After filtering out the low quality reads, 538,387 high quality sequences were obtained varying from 855 to 20,738 sequences per sample, with an average number of 8,157 (SD = 5,303) and a mean sequence length of 358 bp. The number of high quality sequences was distributed among the groups in the following proportion: 24.4% for the untreated mice (n = 15), 22.1% for the vancomycin-treated adults (n = 13), 30.1% for the 4-week-old vancomycin-treated pups (n = 18) and 23.4% for the vancomycin-treated pups as adults (n = 20). Rarefaction curves, and the Shannon and Chao1 indexes revealed a significantly lower bacterial diversity and richness for the vancomycin-treated groups, whereas no difference was found between untreated mice and adult mice that had recovered from neonatal vancomycin treatment.
The vast majority (>99%) of the annotated reads in untreated mice and adult mice that had recovered from neonatal vancomycin treatment were distributed among three bacterial phyla: Firmicutes (Gram positive), Bacteroidetes (Gram negative) and Verrucomicrobia (Gram negative) (Fig. 3). Verrucomicrobia turned out to be the most abundant phylum (>80%) in both groups of vancomycin-treated mice. All reads within this phylum belonged to the genus Akkermansia. The genus Akkermansia consists of a single species, the mucin-degrading Akkermansia muciniphila [29, 30]. The annotation of reads within this genus to A. muciniphila was confirmed by manually comparing sequences retrieved from the dataset against the 16s rRNA gene sequence of the A. muciniphila type strain (ATCC BAA-835T) deposited in the NCBI GenBank database (AY271254). In all tested cases (n = 10) 100% similarity was found.
Pie charts illustrating phylum distribution data from 454 tag sequencing based on 16S rRNA gene PCR-derived amplicons of faeces samples collected from untreated control mice (a), mice that have recovered from neonatal vancomycin treatment (b) and mice treated with vancomycin as adults (c) or as pups (d). Classification into taxa was carried out using RDP’s Pyrosequencing Pipeline (confidence threshold 80%). See text for details. The unlabelled thin light grey section seen next to Firmicutes in an anticlockwise direction represents a small group (<1%) of ‘other bacteria’
Vancomycin treatment of both adults and pups induced a significant decrease in the phyla Firmicutes and Bacteroidetes (Fig. 3). Species in the order Clostridiales represented more than 95% of the Firmicute annotated reads in the untreated mice and several classified and unclassified Ruminococcaceae genera were found in lower frequency in both groups of vancomycin-treated mice compared with the untreated mice (Fig. 4). However, the biggest decrease in Firmicutes during treatment was observed in the Lachnospiraceae family, primarily among unclassified genera. The eradicated Bacteroidetes species were represented by species within the order Bacteroidales, primarily unclassified Porphyromonadacea, but also Barnesiella and Alistipes species (Fig. 4). The gut microbiota harboured several genera belonging to Proteobacteria in the neonatally treated mice; for example Proteus increased from very low levels in untreated mice to almost 7% of all reads during vancomycin treatment (Fig. 4).
Area chart illustrating genus distribution data from 454 tag sequencing based on 16S rRNA gene PCR-derived amplicons of faeces samples collected from untreated control mice (C), mice that have recovered from neonatal vancomycin treatment (R), mice treated with vancomycin as adults (VA) or as pups (VP). Taxon classification was carried out using RDP’s Pyrosequencing Pipeline (confidence threshold 80%). See text for details. Bacteroidetes (red/pink), Firmicutes (blue/purple), Verrucomicrobia (green), and Proteobacteria (yellow/orange) genera are shown. Genera representing less than 0.01% of all reads were filtered out
Early life treatment with vancomycin disrupts intestinal cytokine balance
To elucidate the influence of vancomycin treatment on intestinal immunity, we used flow cytometry to examine the cytokine production in cluster of differentiation (CD)4+ T cells in the small intestinal lamina propria (siLP) and spleen. Neonatally vancomycin-treated mice had a significantly increased frequency of siLP CD4+ T cells producing pro-inflammatory cytokines IFN-γ and TNF-α as adults compared with untreated mice (p < 0.05). The frequency of IL-17 producing CD4+ T cells was also higher, but not significantly so (Fig. 5). In contrast, the proportion of IL-10+CD4+ T cells was the same as in siLP tissue from untreated mice (Fig. 5). To investigate whether this occurred systemically, we analysed the same cytokine profiles in the spleen and found no differences in cytokine levels among the vancomycin-treated mice compared with the untreated mice. Furthermore, treatment of neonate or adult NOD mice with vancomycin did not change the proportion of FOXP3+ Tregs or CD103+ tolerogenic DCs in MLNs, PLNs and spleen.
IFN-γ (a, b), TNF-α (c, d), IL-17 (e, f) and IL-10 (g, h) expression in CD4+ lymphocytes isolated from the entire siLP tissue of untreated control mice (C), mice treated with vancomycin as adults (VA) and mice that have recovered from neonatal vancomycin treatment (R). The lymphocytes were stimulated for 4 h with 50 ng/ml PMA and 750 ng/ml ionomycin in the presence of BD GolgiStop before the cells were harvested and stained intracellularly for cytokines. (a, c, e, g) Error bars represent the SEM. * p < 0.05 compared with untreated mice. (b, d, f, h) Selected data are shown as dot plots gated on CD4+ cells
Discussion
This study addresses the role of gut microbiota in early life as an environmental factor modifying the development of autoimmune diabetes in NOD mice. On treatment of NOD mice with the glycopeptide antimicrobial drug vancomycin from birth until weaning, the cumulative diabetes incidence significantly decreased. In contrast, vancomycin treatment later in life resulted in a non-significant decrease in the cumulative diabetes incidence, which suggests that the impact of intestinal microbial signalling in NOD mice is mainly central to diabetes development at the beginning of life. Previous studies in other rodent models of type 1 diabetes have similarly shown how differences in bacterial composition detected before clinical onset are associated with disease incidence [31, 32]. NOD mice are highly genetically primed toward a diabetogenic immune response [33] and insulitis development is already initiated at the age of 3 to 5 weeks, which might explain why the decrease in cumulative diabetes incidence during vancomycin treatment in adult mice was not as prominent as in the vancomycin-treated pups.
The adult treated mice displayed significantly lower insulitis score and blood glucose levels compared with the other groups. This suggests that diabetes development may also, in fact, be affected in adult life although the difference in cumulative diabetes incidence was not statistically significant compared with the untreated mice. Assessment of insulitic lesions for cell subsets or cytokine profiles might have provided further information on possible qualitative differences between groups, and should help further characterisation of the effects of adult treatment. The lower insulitis score that only occurred in the adult treated group might very well be connected to the ongoing vancomycin treatment, as only this group were receiving vancomycin at the time the mice were killed. Furthermore, some of the mice developed diabetes only shortly before 30 weeks of age and, thus, it is not clear whether the effect of vancomycin delayed or completely prevented diabetes in the mice that obtained protection. However, as the blood glucose values and average insulitis score for the non-diabetic adult treated mice were significantly lower than for the diabetic adult treated mice, this might indicate that the effect of vancomycin would be sustained for some duration.
Interestingly, A. muciniphila was found to be the dominant species during vancomycin treatment. This bacterium depends on mucin as a carbon and nitrogen source and produces short chain fatty acids from mucin fermentation [29]. Furthermore, this species is found in normal healthy humans but is reduced in patients with inflammatory bowel disease [34] and, thus, it has been suggested that it possesses anti-inflammatory properties.
A multitude of microbes in the gastrointestinal tract has co-evolved in a mutualistic symbiosis through time in which, for example, metabolic products of one organism are required by another. Not surprisingly, treatment with the anti-Gram-positive antibiotic vancomycin led to a decreased incidence of several species besides the vancomycin sensitive species. However, in contrast to a loss of Firmicutes (Gram positive) and Bacteroidetes (Gram negative), the phyla Proteobacteria (Gram negative) and Verrucomicrobia (Gram negative) were present with high incidence in vancomycin-treated groups. This is in agreement with another study in which caecal samples from mice treated with vancomycin from birth contained γ-proteobacteria (the class of which Proteus, Citrobacter and Escherichia/Shigella are members) [23].
It may seem paradoxical that robust depletion of microbiota following vancomycin treatment diminished diabetes, on the basis that a lack of microbes should increase autoimmune disease, according to the widely recognised hygiene hypothesis. Particularly in developed countries, intestinal colonisation with many Gram-negative bacteria happens later in life compared with non-developed countries and this has been hypothesised to be associated with a higher incidence of several autoimmune diseases [21, 35]. Today, this concept is gradually being modified toward a more detailed understanding of the different specificities in microbial signalling that prime the immune system. It is more likely to be dysbiosis, rather than a lack of microbes, that contributes to a change in host immune reactions [36]. Even though vancomycin treatment broadly depleted many major species and genera of Gram-positive and Gram-negative microbes, a prominent increase in A. muciniphila and Proteobacteria was found. A potential loss of mucus in the intestines due to the abundance of the mucin-degrading A. muciniphila might have increased the possibility for other Gram-negative bacteria (e.g. Proteobacteria) in the gut to get in contact with intestinal immune cells and stimulate receptors such as TLR-4, which is known to reduce diabetes incidence [37]. It should, however, be noted that the role of A. muciniphila and Proteobacteria in type 1 diabetes has not been established even though the dominance of these bacteria was accompanied by a decrease in type 1 diabetes development.
Tregs have been found with reduced frequency in jejunum biopsies from type 1 diabetic patients. As such, induction of different subsets of Tregs could be a possible commensal-regulated immune pathway involved in protection from diabetes [38]. Nonetheless, we found no differences among Treg and tolerogenic DC populations in either mucosal or systemic compartments after vancomycin treatment. In addition to this, an increase in pro-inflammatory CD4+ T cells in the gut of neonatally treated mice was found. Unlike our data, previous studies have shown a shift in gut homeostasis toward a pro-inflammatory state preceding an increased susceptibility to type 1 diabetes [39, 40]. Furthermore, a recent paper described a study in which pro-inflammatory cytokines were found in excess in the colon of newly weaned NOD mice [41]. Thus, the pro-inflammatory phenotype found in the neonatally treated mice seems not to be involved in diabetes protective mechanisms, as Th1 lymphocytes are rather thought to increase the likelihood of diabetes. Instead, one possible explanation of the altered immune profile could be the high frequency of mucin-degrading A. muciniphila, as a loss of the protective mucus layer may have exposed the intestinal epithelial cells to other microorganisms inducing some degree of inflammation. A. muciniphila has also been shown in a recent study to directly alter mucosal gene expression profiles in the gut; however the modulation of genes involved in immune-regulatory processes was most pronounced in the colon [42]. Ivanov and co-workers [23] have also previously demonstrated how vancomycin treatment of newborn pups increases production of pro-inflammatory cytokines in the small intestine. It is therefore likely that this finding, which is similar to ours, is an additional, but diabetes-independent, effect of early life vancomycin treatment.
During infancy, the gastrointestinal environment endures major developmental changes and, until the entire ecosystem is established, minor modifications in this critical stage can have crucial consequences for normal intestinal immune homeostasis [43]. Later in life, immunity is kept in check by regulatory mechanisms [44, 45] and becomes more resistant to microbial variation. This might also explain why treating mice after the age of 8 weeks made no difference to the mucosal cytokine balance.
In conclusion, our study demonstrates the importance of the early postnatal period as a critical time point for microbiota induced autoimmune diabetes protection and, in particular, that A. muciniphila and Proteobacteria may be of potential significance. It is not clear how intestinal dysbiosis contributes to autoimmune tissue destruction outside the gut, but it is reasonable to assume that the effect is immunological. Clearly, the effects and underlying mechanisms of specific antigenic loads, for example by mono-colonising germ-free NOD mice with A. muciniphila or subsets of different bacterial species, including Proteobacteria, needs to be further investigated.
Abbreviations
- CD:
-
Cluster of differentiation
- DC:
-
Dendritic cell
- DGGE:
-
Denaturing gradient gel electrophoresis
- FOXP3:
-
Forkhead box P3
- MLN:
-
Mesenteric lymph node
- PCA:
-
Principal component analysis
- PLN:
-
Pancreatic lymph node
- PMA:
-
Phorbol 12-myristate 13-acetate
- RDP:
-
Ribosomal Database Project
- rRNA:
-
Ribosomal RNA
- siLP:
-
Small intestinal lamina propria
- Th:
-
T helper
- TLR:
-
Toll-like receptor
- Treg:
-
Regulatory T cell
References
Fujita T, Yui R, Kusumoto Y, Serizawa Y, Makino S, Tochino Y (1982) Lymphocytic insulitis in a nonobese diabetic (NOD) strain of mice—an immunohistochemical and electron-microscope investigation. Biomedical Research-Tokyo 3:429–443
Vaarala O, Atkinson MA, Neu J (2008) The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 57:2555–2562
Qin JJ, Li RQ, Raes J et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65
Atarashi K, Tanoue T, Shima T et al (2011) Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341
Round JL, Mazmanian SK (2010) Inducible Foxp(3+) regulatory T cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA 107:12204–12209
Ivanov II, Atarashi K, Manel N et al (2009) Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498
Honda K, Takeda K (2009) Regulatory mechanisms of immune responses to intestinal bacteria. Mucosal Immunol 2:187–196
Ishikawa H, Tanaka K, Maeda Y et al (2008) Effect of intestinal microbiota on the induction of regulatory CD25+ CD4+ T cells. Clin Exp Immunol 153:127–135
Min B, Thornton A, Caucheteux SM et al (2007) Gut flora antigens are not important in the maintenance of regulatory T cell heterogeneity and homeostasis. Eur J Immunol 37:1916–1923
Round JL, O’Connell RM, Mazmanian SK (2010) Coordination of tolerogenic immune responses by the commensal microbiota. J Autoimmun 34:J220–J225
Pozzilli P, Signore A, Williams AJK, Beales PE (1993) NOD mouse colonies around the world—recent facts and figures. Immunol Today 14:193–196
Alam C, Bittoun E, Bhagwat D et al (2011) Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia 54:1398–1406
Wen L, Ley RE, Volchkov PY et al (2008) Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455:1109–1113
Buschard K, Pedersen C, Hansen SV, Hageman I, Aaen K, Bendtzen K (1992) Anti-diabetogenic effect of fusidic acid in diabetes prone Bb rats. Autoimmunity 14:101–104
Hansen AK, Ling F, Kaas A, Funda DP, Farlov H, Buschard K (2006) Diabetes preventive gluten-free diet decreases the number of caecal bacteria in non-obese diabetic mice. Diabetes Metab Res Rev 22:220–225
Karlsson MR, Kahu H, Hanson LA, Telemo E, Dahlgren UI (1999) Neonatal colonization of rats induces immunological tolerance to bacterial antigens. Eur J Immunol 29:109–118
Mowat AM (1987) The regulation of immune-responses to dietary-protein antigens. Immunol Today 8:93–98
Mowat AM (1999) Basic mechanisms and clinical implications of oral tolerance. Curr Opin Gastroenterol 15:546–556
Brugman S, Klatter FA, Visser J, Bos NA, Elias D, Rozing J (2004) Neonatal oral administration of DiaPep277, combined with hydrolysed casein diet, protects against Type 1 diabetes in BB-DP rats. An experimental study. Diabetologia 47:1331–1333
Scott FW, Rowsell P, Wang GS, Burghardt K, Kolb H, Flohe S (2002) Oral exposure to diabetes-promoting food or immunomodulators in neonates alters gut cytokines and diabetes. Diabetes 51:73–78
Bach JF (2002) The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med 347:911–920
Wills-Karp M, Santeliz J, Karp CL (2001) The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol 1:69–75
Ivanov II, Frutos RL, Manel N et al (2008) Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4:337–349
Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK (2010) Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp Med 60:336–347
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679
Holmsgaard PN, Hede CR, Poulsen PHB, Al-Soud WA, Hansen LH, Sørensen SJ (2011) Bias in bacterial diversity as a result of Nycodenz extraction from bulk soil. Soil Biol Biochem 43:2152–2159
Larsen N, Vogensen FK, van den Berg FW et al (2010) Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085
Zoetendal EG, Rajilic-Stojanovic M, de Vos WM (2008) High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57:1605–1615
Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM (2008) The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol 74:1646–1648
Hedlund BP, Gosink JJ, Staley JT (1997) Verrucomicrobia div. nov., a new division of the bacteria containing three new species of Prosthecobacter. Antonie van Leeuwenhoek 72:29–38
Brugman S, Klatter FA, Visser JTJ et al (2006) Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 49:2105–2108
Roesch LFW, Lorca GL, Casella G et al (2009) Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J 3:536–548
Wicker LS, Miller BJ, Coker LZ et al (1987) Genetic-control of diabetes and insulitis in the nonobese diabetic (NOD) mouse. J Exp Med 165:1639–1654
Png CW, Linden SK, Gilshenan KS et al (2010) Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105:2420–2428
Okada H, Kuhn C, Feillet H, Bach JF (2010) The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin Exp Immunol 160:1–9
Reading NC, Kasper DL (2011) The starting lineup: key microbial players in intestinal immunity and homeostasis. Front Microbiol 2:148
Wang ZJ, Zhang FM, Wang LS, Yao YW, Zhao Q, Gao X (2009) Lipopolysaccharides can protect mesenchymal stem cells (MSCs) from oxidative stress-induced apoptosis and enhance proliferation of MSCs via Toll-like receptor(TLR)-4 and PI3K/Akt. Cell Biol Int 33:665–674
Badami E, Sorini C, Coccia M et al (2011) defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes 60:2120–2124
Savilahti E, Ormala T, Saukkonen T et al (1999) Jejuna of patients with insulin-dependent diabetes mellitus (IDDM) show signs of immune activation. Clin Exp Immunol 116:70–77
Westerholm-Ormio M, Vaarala O, Pihkala P, Ilonen J, Savilahti E (2003) Immunologic activity in the small intestinal mucosa of pediatric patients with type 1 diabetes. Diabetes 52:2287–2295
Alam C, Valkonen S, Palagani V, Jalava J, Eerola E, Hanninen A (2010) Inflammatory tendencies and overproduction of IL-17 in the colon of young NOD mice are counteracted with diet change. Diabetes 59:2237–2246
Derrien M, van Baarlen P, Hooiveld G, Norin E, Muller M, de Vos WM (2011) Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol 2:166
Sudo N, Yu XN, Aiba Y et al (2002) An oral introduction of intestinal bacteria prevents the development of a long-term Th2-skewed immunological memory induced by neonatal antibiotic treatment in mice. Clin Exp Allergy 32:1112–1116
Cebra JJ (1999) Influences of microbiota on intestinal immune system development. Am J Clin Nutr 69:1046S–1051S
Tlaskalova-Hogenova H, Stepankova R, Hudcovic T et al (2004) Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett 93:97–108
Acknowledgments
H. Farlov, Faculty of Life Sciences, University of Copenhagen, Denmark, is kindly acknowledged for taking care of the mice.
Funding
This work was supported by grants from Arkitekt Holger Hjortenberg og hustru Dagmar Hjortenbergs fond, the Center for Applied Laboratory Animal Research (www.calar.dk), and the Chemometric Analysis Centre (www.chance.life.ku.dk). It was carried out as part of the research programme of the UNIK: Food, Fitness & Pharma for Health and Disease (www.foodfitnesspharma.ku.dk). The UNIK project is supported by the Danish Ministry of Science, Technology and Innovation.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
CHFH and AKH were responsible for the conception of the study and designed the experiments. CHFH, LK, KB, FKV, DSN, SJS, and LHH analysed and interpreted data. CHFH, LK, and AKH drafted the article and KB, FKV, DSN, SJS, and LHH revised it critically for important intellectual content. All authors approved the final version.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Table 1
(PDF 10 kb)
Rights and permissions
About this article
Cite this article
Hansen, C.H.F., Krych, L., Nielsen, D.S. et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 55, 2285–2294 (2012). https://doi.org/10.1007/s00125-012-2564-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-012-2564-7