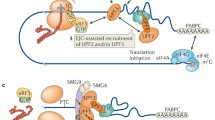
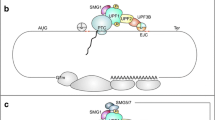
Abstract
Nonsense-mediated decay is well known by the lucid definition of being a RNA surveillance mechanism that ensures the speedy degradation of mRNAs containing premature translation termination codons. However, as we review here, NMD is far from being a simple quality control mechanism; it also regulates the stability of many wild-type transcripts. We summarise the abundance of research that has characterised each of the NMD factors and present a unified model for the recognition of NMD substrates. The contentious issue of how and where NMD occurs is also discussed, particularly with regard to P-bodies and SMG6-driven endonucleolytic degradation. In recent years, the discovery of additional functions played by several of the NMD factors has further complicated the picture. Therefore, we also review the reported roles of UPF1, SMG1 and SMG6 in other cellular processes.



Similar content being viewed by others
References
Losson R, Lacroute F (1979) Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA 76:5134–5137
Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J (1981) Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell 27:543–553
Culbertson MR, Leeds PF (2003) Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev 13:207–214
Stalder L, Muhlemann O (2008) The meaning of nonsense. Trends Cell Biol 18:315–321
Li S, Wilkinson MF (1998) Nonsense surveillance in lymphocytes? Immunity 8:135–141
Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT (2008) NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22:1381–1396
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415
Kim H, Klein R, Majewski J, Ott J (2004) Estimating rates of alternative splicing in mammals and invertebrates. Nat Genet 36:915–916; author reply 916–917
Lewis BP, Green RE, Brenner SE (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA 100:189–192
Isken O, Maquat LE (2008) The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet 9:699–712
McGlincy NJ, Smith CW (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33:385–393
Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ (2006) Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev 20:153–158
Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, Toyoda T, Shinozaki K, Seki M (2009) Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci USA 106:2453–2458
Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36:1073–1078
Mitrovich QM, Anderson P (2005) mRNA surveillance of expressed pseudogenes in C. elegans. Curr Biol 15:963–967
Rehwinkel J, Raes J, Izaurralde E (2006) Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem Sci 31:639–646
Lelivelt MJ, Culbertson MR (1999) Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol 19:6710–6719
He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A (2003) Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12:1439–1452
Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E (2005) Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11:1530–1544
Wittmann J, Hol EM, Jack HM (2006) hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26:1272–1287
Rodriguez-Gabriel MA, Watt S, Bahler J, Russell P (2006) Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol Cell Biol 26:6347–6356
Moriarty PM, Reddy CC, Maquat LE (1998) Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol 18:2932–2939
Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE (2004) Nonsense-mediated decay approaches the clinic. Nat Genet 36:801–808
Khajavi M, Inoue K, Lupski JR (2006) Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet 14:1074–1081
Kuzmiak HA, Maquat LE (2006) Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med 12:306–316
Kerr TP, Sewry CA, Robb SA, Roberts RG (2001) Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum Genet 109:402–407
Hall GW, Thein S (1994) Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood 83:2031–2037
Thein SL, Hesketh C, Taylor P, Temperley IJ, Hutchinson RM, Old JM, Wood WG, Clegg JB, Weatherall DJ (1990) Molecular basis for dominantly inherited inclusion body beta-thalassemia. Proc Natl Acad Sci USA 87:3924–3928
Dietz HC, McIntosh I, Sakai LY, Corson GM, Chalberg SC, Pyeritz RE, Francomano CA (1993) Four novel FBN1 mutations: significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics 17:468–475
Salvatore F, Scudiero O, Castaldo G (2002) Genotype-phenotype correlation in cystic fibrosis: the role of modifier genes. Am J Med Genet 111:88–95
Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile JF, Newport M, Levin M, Blanche S, Seboun E, Fischer A, Casanova JL (1996) Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N Engl J Med 335:1956–1961
Jouanguy E, Lamhamedi-Cherradi S, Lammas D, Dorman SE, Fondaneche MC, Dupuis S, Doffinger R, Altare F, Girdlestone J, Emile JF, Ducoulombier H, Edgar D, Clarke J, Oxelius VA, Brai M, Novelli V, Heyne K, Fischer A, Holland SM, Kumararatne DS, Schreiber RD, Casanova JL (1999) A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat Genet 21:370–378
Cardinali M, Kratochvil FJ, Ensley JF, Robbins KC, Yeudall WA (1997) Functional characterization in vivo of mutant p53 molecules derived from squamous cell carcinomas of the head and neck. Mol Carcinog 18:78–88
Englert C, Vidal M, Maheswaran S, Ge Y, Ezzell RM, Isselbacher KJ, Haber DA (1995) Truncated WT1 mutants alter the subnuclear localization of the wild-type protein. Proc Natl Acad Sci USA 92:11960–11964
Reddy JC, Morris JC, Wang J, English MA, Haber DA, Shi Y, Licht JD (1995) WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J Biol Chem 270:10878–10884
Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S (2002) The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet 11:2805–2814
Bloethner S, Mould A, Stark M, Hayward NK (2008) Identification of ARHGEF17, DENND2D, FGFR3, and RB1 mutations in melanoma by inhibition of nonsense-mediated mRNA decay. Genes Chromosomes Cancer 47:1076–1085
Ivanov I, Lo KC, Hawthorn L, Cowell JK, Ionov Y (2007) Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene 26:2873–2884
Tarpey PS, Lucy Raymond F, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O’Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Hills K, Jones D, Mironenko T, Perry J, Varian J, West S, Widaa S, Teague J, Dicks E, Butler A, Menzies A, Richardson D, Jenkinson A, Shepherd R, Raine K, Moon J, Luo Y, Parnau J, Bhat SS, Gardner A, Corbett M, Brooks D, Thomas P, Parkinson-Lawrence E, Porteous ME, Warner JP, Sanderson T, Pearson P, Simensen RJ, Skinner C, Hoganson G, Superneau D, Wooster R, Bobrow M, Turner G, Stevenson RE, Schwartz CE, Andrew Futreal P, Srivastava AK, Stratton MR, Gecz J (2007) Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 39:1127–1133
Kerem E (2004) Pharmacologic therapy for stop mutations: how much CFTR activity is enough? Curr Opin Pulm Med 10:547–552
Ramalho AS, Beck S, Meyer M, Penque D, Cutting GR, Amaral MD (2002) Five percent of normal cystic fibrosis transmembrane conductance regulator mRNA ameliorates the severity of pulmonary disease in cystic fibrosis. Am J Respir Cell Mol Biol 27:619–627
Stephenson J (2001) Antibiotics show promise as therapy for genetic disorders. JAMA 285:2067–2068
Clancy JP, Bebok Z, Ruiz F, King C, Jones J, Walker L, Greer H, Hong J, Wing L, Macaluso M, Lyrene R, Sorscher EJ, Bedwell DM (2001) Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am J Respir Crit Care Med 163:1683–1692
Wilschanski M, Famini C, Blau H, Rivlin J, Augarten A, Avital A, Kerem B, Kerem E (2000) A pilot study of the effect of gentamicin on nasal potential difference measurements in cystic fibrosis patients carrying stop mutations. Am J Respir Crit Care Med 161:860–865
Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, Wilde RG, Karp G, Takasugi J, Chen G, Jones S, Ren H, Moon YC, Corson D, Turpoff AA, Campbell JA, Conn MM, Khan A, Almstead NG, Hedrick J, Mollin A, Risher N, Weetall M, Yeh S, Branstrom AA, Colacino JM, Babiak J, Ju WD, Hirawat S, Northcutt VJ, Miller LL, Spatrick P, He F, Kawana M, Feng H, Jacobson A, Peltz SW, Sweeney HL (2007) PTC124 targets genetic disorders caused by nonsense mutations. Nature 447:87–91
Kerem E, Hirawat S, Armoni S, Yaakov Y, Shoseyov D, Cohen M, Nissim-Rafinia M, Blau H, Rivlin J, Aviram M, Elfring GL, Northcutt VJ, Miller LL, Kerem B, Wilschanski M (2008) Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet 372:719–727
Culbertson MR, Underbrink KM, Fink GR (1980) Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics 95:833–853
Leeds P, Peltz SW, Jacobson A, Culbertson MR (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5:2303–2314
Leeds P, Wood JM, Lee BS, Culbertson MR (1992) Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol 12:2165–2177
He F, Jacobson A (1995) Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev 9:437–454
Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P (1989) A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123:301–313
Pulak R, Anderson P (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 7:1885–1897
Cali BM, Kuchma SL, Latham J, Anderson P (1999) smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151:605–616
Applequist SE, Selg M, Raman C, Jack HM (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res 25:814–821
Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP (2001) Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J Biol Chem 276:22709–22714
Lykke-Andersen J, Shu MD, Steitz JA (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121–1131
Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol 20:8944–8957
Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S (2003) Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12:1187–1200
Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ Jr, Dietz HC (1996) Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci USA 93:10928–10932
Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21:209–223
Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15:2215–2228
Longman D, Plasterk RH, Johnstone IL, Caceres JF (2007) Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 21:1075–1085
Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, Kurata R, Hirano H, Anderson P, Ohno S (2009) SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23:1091–1105
Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E (2003) Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 22:3960–3970
Luke B, Azzalin CM, Hug N, Deplazes A, Peter M, Lingner J (2007) Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res 35:7688–7697
Page MF, Carr B, Anders KR, Grimson A, Anderson P (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19:5943–5951
Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6:1226–1235
Czaplinski K, Weng Y, Hagan KW, Peltz SW (1995) Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1:610–623
Cheng Z, Muhlrad D, Lim MK, Parker R, Song H (2007) Structural and functional insights into the human Upf1 helicase core. EMBO J 26:253–264
Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S (2009) Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J 28:2293–2306
Kadlec J, Guilligay D, Ravelli RB, Cusack S (2006) Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 12:1817–1824
Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12:1665–1677
Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27:736–747
Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20:355–367
Grimson A, O’Connor S, Newman CL, Anderson P (2004) SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol Cell Biol 24:7483–7490
Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA 2nd, Wilkinson MF (2007) An alternative branch of the nonsense-mediated decay pathway. EMBO J 26:1820–1830
Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE (2005) Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20:65–75
Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE (2009) The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol 7:e1000120
Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ (2008) Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28:4320–4330
Anders KR, Grimson A, Anderson P (2003) SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22:641–650
Chiu SY, Serin G, Ohara O, Maquat LE (2003) Characterization of human Smg5/7a: A protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9:77–87
Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, Gonzalez CI (2006) Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol Cell Biol 26:3390–3400
He F, Brown AH, Jacobson A (1997) Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol 17:1580–1594
Weng Y, Czaplinski K, Peltz SW (1996) Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol 16:5491–5506
Kadlec J, Izaurralde E, Cusack S (2004) The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11:330–337
Schell T, Kocher T, Wilm M, Seraphin B, Kulozik AE, Hentze MW (2003) Complexes between the nonsense-mediated mRNA decay pathway factor human upf1 (up-frameshift protein 1) and essential nonsense-mediated mRNA decay factors in HeLa cells. Biochem J 373:775–783
Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF (2009) A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol 16:747–753
Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH (2006) Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. Rna 12:1015–1022
Le Hir H, Gatfield D, Izaurralde E, Moore MJ (2001) The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20:4987–4997
Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR (2006) Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313:1968–1972
Bono F, Ebert J, Lorentzen E, Conti E (2006) The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126:713–725
Chamieh H, Ballut L, Bonneau F, Le Hir H (2008) NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15:85–93
Kim VN, Kataoka N, Dreyfuss G (2001) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science 293:1832–1836
Yamashita A, Kashima I, Ohno S (2005) The role of SMG-1 in nonsense-mediated mRNA decay. Biochim Biophys Acta 1754:305–315
D’Andrea LD, Regan L (2003) TPR proteins: the versatile helix. Trends Biochem Sci 28:655–662
Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14–3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17:537–547
Unterholzner L, Izaurralde E (2004) SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16:587–596
Clissold PM, Ponting CP (2000) PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol 10:R888–R890
Glavan F, Behm-Ansmant I, Izaurralde E, Conti E (2006) Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25:5117–5125
Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH (2009) SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16:49–55
Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E (2008) SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14:2609–2617
Mendell JT, ap Rhys CM, Dietz HC (2002) Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298:419–422
Azzalin CM, Lingner J (2006) The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16:433–439
Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318:798–801
Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejeune F (2007) Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol 178:1145–1160
Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT (2004) The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell 14:585–598
Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW (2009) Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21:2045–2057
Hilleren P, Parker R (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 5:711–719
Amrani N, Sachs MS, Jacobson A (2006) Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7:415–425
Muhlemann O, Eberle AB, Stalder L, Zamudio Orozco R (2008) Recognition and elimination of nonsense mRNA. Biochim Biophys Acta 1779:538–549
Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 14:4365–4373
Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 14:4065–4072
Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, Justesen J, Kisselev L (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372:701–703
Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, Zhouravleva G (2002) Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol Cell 94:205–216
Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T (1999) The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3’-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem 274:16677–16680
Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K (2001) Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci USA 98:4409–4413
Mangus DA, Evans MC, Jacobson A (2003) Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4:223
Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432:112–118
Singh G, Rebbapragada I, Lykke-Andersen J (2008) A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6:e111
Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E (2007) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26:1591–1601
Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O (2008) Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6:e92
Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L (2008) Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14:563–576
Kebaara BW, Atkin AL (2009) Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res 37:2771–2778
Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D (2008) Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 27:1585–1595
Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D (2006) Both introns and long 3’-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 34:6147–6157
Muhlrad D, Parker R (1999) Aberrant mRNAs with extended 3 ‘UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5:1299–1307
Peltz SW, Brown AH, Jacobson A (1993) mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev 7:1737–1754
Ruiz-Echevarria MJ, Gonzalez CI, Peltz SW (1998) Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J 17:575–589
Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19:6860–6869
Le Hir H, Moore MJ, Maquat LE (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev 14:1098–1108
Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17:3484–3494
Zhang J, Sun X, Qian Y, La Duca JP, Maquat LE (1998) At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol 18:5272–5283
Zhang J, Sun X, Qian Y, Maquat LE (1998) Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4:801–815
Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW (2009) Disassembly of exon junction complexes by PYM. Cell 137:536–548
Buzina A, Shulman MJ (1999) Infrequent translation of a nonsense codon is sufficient to decrease mRNA level. Mol Biol Cell 10:515–524
Inacio A, Silva AL, Pinto J, Ji X, Morgado A, Almeida F, Faustino P, Lavinha J, Liebhaber SA, Romao L (2004) Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J Biol Chem 279:32170–32180
Romao L, Inacio A, Santos S, Avila M, Faustino P, Pacheco P, Lavinha J (2000) Nonsense mutations in the human beta-globin gene lead to unexpected levels of cytoplasmic mRNA accumulation. Blood 96:2895–2901
Zhang J, Maquat LE (1997) Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J 16:826–833
Tarun SZ Jr, Sachs AB (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15:7168–7177
Tarun SZ Jr, Wells SE, Deardorff JA, Sachs AB (1997) Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA 94:9046–9051
Silva AL, Romao L (2009) The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS Lett 583:499–505
Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation. mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106:607–617
Chiu SY, Lejeune F, Ranganathan AC, Maquat LE (2004) The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev 18:745–754
Hosoda N, Kim YK, Lejeune F, Maquat LE (2005) CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 12:893–901
Gao Q, Das B, Sherman F, Maquat LE (2005) Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc Natl Acad Sci USA 102:4258–4263
Maderazo AB, Belk JP, He F, Jacobson A (2003) Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol Cell Biol 23:842–851
Kuperwasser N, Brogna S, Dower K, Rosbash M (2004) Nonsense-mediated decay does not occur within the yeast nucleus. RNA 10:1907–1915
Brogna S, Wen J (2009) Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol 16:107–113
Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8:113–126
Meyer S, Temme C, Wahle E (2004) Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol 39:197–216
Coller J, Parker R (2004) Eukaryotic mRNA decapping. Annu Rev Biochem 73:861–890
Parker R, Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11:121–127
Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB (2005) Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12:1054–1063
Schmid M, Jensen TH (2008) The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci 33:501–510
Cao D, Parker R (2003) Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113:533–545
Muhlrad D, Parker R (1994) Premature translational termination triggers mRNA decapping. Nature 370:578–581
Mitchell P, Tollervey D (2003) An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′->5′ degradation. Molecular Cell 11:1405–1413
Gatfield D, Izaurralde E (2004) Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429:575–578
Couttet P, Grange T (2004) Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res 32:488–494
Lejeune F, Li X, Maquat LE (2003) Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12:675–687
Chen CY, Shyu AB (2003) Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol 23:4805–4813
Cho H, Kim KM, Kim YK (2009) Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 33:75–86
Franks TM, Lykke-Andersen J (2008) The control of mRNA decapping and P-body formation. Mol Cell 32:605–615
Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22
Parker R, Sheth U (2007) P bodies and the control of mRNA translation and degradation. Mol Cell 25:635–646
Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB (2008) Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 182:89–101
Sheth U, Parker R (2006) Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125:1095–1109
Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E (2007) P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27:3970–3981
Stalder L, Muhlemann O (2009) Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA 15:1265–1273
Metzstein MM, Krasnow MA (2006) Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet 2:e180
Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E (2009) Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol 29:3517–3528
Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 10:99–105
Yoine M, Nishii T, Nakamura K (2006) Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol 47:572–580
Baron C, Heider J, Bock A (1993) Interaction of translation factor SELB with the formate dehydrogenase H selenopolypeptide mRNA. Proc Natl Acad Sci USA 90:4181–4185
Caban K, Copeland PR (2006) Size matters: a view of selenocysteine incorporation from the ribosome. Cell Mol Life Sci 63:73–81
Donovan J, Caban K, Ranaweera R, Gonzalez-Flores JN, Copeland PR (2008) A novel protein domain induces high affinity selenocysteine insertion sequence binding and elongation factor recruitment. J Biol Chem 283:35129–35139
Gupta M, Copeland PR (2007) Functional analysis of the interplay between translation termination, selenocysteine codon context, and selenocysteine insertion sequence-binding protein 2. J Biol Chem 282:36797–36807
Wen W, Weiss SL, Sunde RA (1998) UGA codon position affects the efficiency of selenocysteine incorporation into glutathione peroxidase-1. J Biol Chem 273:28533–28541
Howard MT, Moyle MW, Aggarwal G, Carlson BA, Anderson CB (2007) A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA 13:912–920
Sun X, Li X, Moriarty PM, Henics T, LaDuca JP, Maquat LE (2001) Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol Biol Cell 12:1009–1017
Bermano G, Nicol F, Dyer JA, Sunde RA, Beckett GJ, Arthur JR, Hesketh JE (1995) Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem J 311:425–430
St Johnston D, Beuchle D, Nusslein-Volhard C (1991) Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66:51–63
St Johnston D (2001) The beginning of the end. EMBO J 20:6169–6179
Ferrandon D, Elphick L, Nusslein-Volhard C, St Johnston D (1994) Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79:1221–1232
Kim-Ha J, Kerr K, Macdonald PM (1995) Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81:403–412
Marion RM, Fortes P, Beloso A, Dotti C, Ortin J (1999) A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol 19:2212–2219
Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L (1999) Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol 19:2220–2230
Kim YK, Furic L, Desgroseillers L, Maquat LE (2005) Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120:195–208
Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE (2007) Staufen1 regulates diverse classes of mammalian transcripts. EMBO J 26:2670–2681
Furic L, Maher-Laporte M, DesGroseillers L (2008) A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA 14:324–335
Monshausen M, Gehring NH, Kosik KS (2004) The mammalian RNA-binding protein Staufen2 links nuclear and cytoplasmic RNA processing pathways in neurons. Neuromolecular Med 6:127–144
Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L (2005) Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res 33:4797–4812
Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE (2009) SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 23:54–66
Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ (2008) Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14:914–927
Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ (2004) Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol 24:2637–2648
Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA (2000) The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol 74:5441–5451
Falcon AM, Fortes P, Marion RM, Beloso A, Ortin J (1999) Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res 27:2241–2247
Cech TR (2004) Beginning to understand the end of the chromosome. Cell 116:273–279
Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73:177–208
Dahlseid JN, Lew-Smith J, Lelivelt MJ, Enomoto S, Ford A, Desruisseaux M, McClellan M, Lue N, Culbertson MR, Berman J (2003) mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryotic Cell 2:134–142
Lew JE, Enomoto S, Berman J (1998) Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol Cell Biol 18:6121–6130
Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J (2003) A human homolog of yeast est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13:568–574
Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L (2003) Functional conservation of the telomerase protein Est1p in humans. Curr Biol 13:698–704
Redon S, Reichenbach P, Lingner J (2007) Protein RNA and protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res 35:7011–7022
Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10:228–236
Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J (2008) The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32:465–477
Chawla R, Azzalin CM (2008) The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res 122:194–201
Carastro LM, Tan CK, Selg M, Jack HM, So AG, Downey KM (2002) Identification of delta helicase as the bovine homolog of HUPF1: demonstration of an interaction with the third subunit of DNA polymerase delta. Nucleic Acids Res 30:2232–2243
Kaygun H, Marzluff WF (2005) Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol 25:6879–6888
Marzluff WF (2005) Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol 17:274–280
Osley MA (1991) The regulation of histone synthesis in the cell cycle. Annu Rev Biochem 60:827–861
Marzluff WF, Wagner EJ, Duronio RJ (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9:843–854
Nicholson P, Muller B (2008) Post-transcriptional control of animal histone gene expression–not so different after all. Mol Biosyst 4:721–725
Kaygun H, Marzluff WF (2005) Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12:794–800
Muller B, Blackburn J, Feijoo C, Zhao X, Smythe C (2007) DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J Cell Biol 179:1385–1398
Su C, Gao G, Schneider S, Helt C, Weiss C, O’Reilly MA, Bohmann D, Zhao J (2004) DNA damage induces downregulation of histone gene expression through the G(1) checkpoint pathway. EMBO J 23:1133–1143
Mullen TE, Marzluff WF (2008) Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22:50–65
Wilusz CJ, Wilusz J (2008) New ways to meet your (3’) end oligouridylation as a step on the path to destruction. Genes Dev 22:1–7
Zhou BB, Elledge SJ (2000) The DNA damage response: putting checkpoints in perspective. Nature 408:433–439
Morita T, Yamashita A, Kashima I, Ogata K, Ishiura S, Ohno S (2007) Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J Biol Chem 282:7799–7808
Abraham RT (2004) PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 3:883–887
Gehen SC, Staversky RJ, Bambara RA, Keng PC, O’Reilly MA (2008) hSMG-1 and ATM sequentially and independently regulate the G1 checkpoint during oxidative stress. Oncogene 27:4065–4074
Masse I, Molin L, Mouchiroud L, Vanhems P, Palladino F, Billaud M, Solari F (2008) A novel role for the SMG-1 kinase in lifespan and oxidative stress resistance in Caenorhabditis elegans. PLoS One 3:e3354
Oliveira V, Romanow WJ, Geisen C, Otterness DM, Mercurio F, Wang HG, Dalton WS, Abraham RT (2008) A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J Biol Chem 283:13174–13184
Pal M, Ishigaki Y, Nagy E, Maquat LE (2001) Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7:5–15
Wang W, Czaplinski K, Rao Y, Peltz SW (2001) The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20:880–890
Azzalin CM, Lingner J (2006) The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle 5:1496–1498
Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE (2003) Y14 and hUpf3b form an NMD-activating complex. Molecular Cell 11:939–949
He F, Brown AH, Jacobson A (1996) Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA 2:153–170
Acknowledgments
The research of the authors is supported by grants from the European Research Council, the Swiss National Science Foundation, the Novartis Foundation for Biomedical Research, the Helmut Horten Foundation and also by the Kanton Bern. O.M. is a fellow of the Max Cloëtta Foundation and R.Z.O. is supported by a fellowship from CONACYT México.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nicholson, P., Yepiskoposyan, H., Metze, S. et al. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 67, 677–700 (2010). https://doi.org/10.1007/s00018-009-0177-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0177-1