Abstract
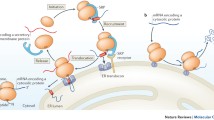
The partitioning of secretory and membrane protein-encoding mRNAs to the endoplasmic reticulum (ER), and their translation on ER-associated ribosomes, governs access to the secretory/exocytic pathways of the cell. As mRNAs encoding secretory and membrane proteins comprise approximately 30% of the transcriptome, the localization of mRNAs to the ER represents an extraordinarily prominent, ubiquitous, and yet poorly understood RNA localization phenomenon.
The partitioning of mRNAs to the ER is generally thought to be achieved by the signal recognition particle (SRP) pathway. In this pathway, mRNA localization to the ER is determined by the translation product – translation yields an N-terminal signal sequence or a topogenic signal that is recognized by the SRP and the resulting mRNA–ribosome–SRP complex is then recruited to the ER membrane. Recent studies have demonstrated that mRNAs can be localized to the ER via a signal sequence and/or translation-independent pathway(s) and that discrete sets of cytosolic protein-encoding mRNAs are enriched on the ER membrane, though they lack an encoded signal sequence. These key findings reopen investigations into the mechanism(s) that govern mRNA localization to the ER.
In this contribution, we describe two independent methods that can be utilized to study this important and poorly understood aspect of eukaryotic cell biology. These methods comprise two independent means of fractionating tissue culture cells to yield free/cytosolic polyribosomes and ER membrane-bound polyribosomes. Detailed methods for the fractionation and characterization of the two polyribosome pools are provided.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Stevens, T. J., and Arkin, I. T. (2000) Do more complex organisms have a greater proportion of membrane proteins in their genomes?, Proteins 39, 417– 420.
Walter, P., Ibrahimi, I., and Blobel, G. (1981) Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein, J Cell Biol 91, 545 – 550.
Walter, P., and Blobel, G. (1981) Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein, J Cell Biol 91, 551– 556.
Walter, P., and Blobel, G. (1981) Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes, J Cell Biol 91, 557– 561.
Lingappa, V. R., and Blobel, G. (1980) Early events in the biosynthesis of secretory and membrane proteins: the signal hypothesis, Recent Prog Horm Res 36, 451– 475.
Lerner, R. S., Seiser, R. M., Zheng, T., Lager, P. J., Reedy, M. C., Keene, J. D., and Nicchitta, C. V. (2003) Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes, RNA 9, 1123–1137.
Diehn, M., Eisen, M. B., Botstein, D., and Brown, P. O. (2000) Large-scale identification of secreted and membrane-associated gene products using DNA microarrays, Nat Genet 25, 58 – 62.
Mueckler, M. M., and Pitot, H. C. (1981) Structure and function of rat liver polysome populations. I. Complexity, frequency distribution, and degree of uniqueness of free and membrane-bound polysomal polyadenylate-containing RNA populations, J Cell Biol 90, 495 – 506.
Mutka, S. C., and Walter, P. (2001) Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway, Mol Biol Cell 12, 577– 588.
Liu, L., Liang, X. H., Uliel, S., Unger, R., Ullu, E., and Michaeli, S. (2002) RNA interference of signal peptide-binding protein SRP54 elicits deleterious effects and protein sorting defects in trypanosomes, J Biol Chem 277, 47348 – 47357.
Ren, Y. G., Wagner, K. W., Knee, D. A., Aza-Blanc, P., Nasoff, M., and Deveraux, Q. L. (2004) Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle, Mol Biol Cell 15, 5064 –5074.
Pyhtila, B., Zheng, T., Lager, P. J., Keene, J. D., Reedy, M. C., and Nicchitta, C. V. (2008) Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum, RNA 14, 445 – 453.
Diehn, M., Bhattacharya, R., Botstein, D., and Brown, P. O. (2006) Genome-scale identification of membrane-associated human mRNAs, PLoS Genet 2, e11.
Aragon, T., van Anken, E., Pincus, D., Serafimova, I. M., Korennykh, A. V., Rubio, C. A., and Walter, P. (2008) Messenger RNA targeting to endoplasmic reticulum stress signalling sites, Nature.
Claude, A. (1946) Fractionation of mammalian liver cells by differential centrifugation: II. Experimental procedures and results, Journal of Experimental Medicine 84, 61– 89.
Duve, C. (1975) Exploring cells with a centrifuge, Science 189, 186 –194.
Palacios, I. M., and St Johnston, D. (2001) Getting the message across: the intracellular localization of mRNAs in higher eukaryotes, Annu Rev Cell Dev Biol 17, 569– 614.
Paquin, N., and Chartrand, P. (2008) Local regulation of mRNA translation: new insights from the bud, Trends Cell Biol 18, 105 –111.
Sambrook J, F. E. F., and Maniatis T (2001) Molecular Cloning: A laboratory manual, Cold Spring Harbor Press, Cold Spring Harbor, New York, USA.
Acknowledgements
The authors thank Angela Jockheck-Clark for critical reading of the manuscript. We thank Tianli Zheng and other present and past members of the Nicchitta laboratory for their contribution to the development of the techniques described in this article. We also wish to thank Mayya Shveygert and the Gromeier lab for providing GAPDH and H3 antibodies. This work was supported by NIH grant GM-077382 to CVN.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer Science+Business Media, LLC
About this protocol
Cite this protocol
Jagannathan, S., Nwosu, C., Nicchitta, C.V. (2011). Analyzing mRNA Localization to the Endoplasmic Reticulum via Cell Fractionation. In: Gerst, J. (eds) RNA Detection and Visualization. Methods in Molecular Biology, vol 714. Humana Press, Totowa, NJ. https://doi.org/10.1007/978-1-61779-005-8_19
Download citation
DOI: https://doi.org/10.1007/978-1-61779-005-8_19
Published:
Publisher Name: Humana Press, Totowa, NJ
Print ISBN: 978-1-61779-004-1
Online ISBN: 978-1-61779-005-8
eBook Packages: Springer Protocols