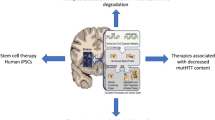
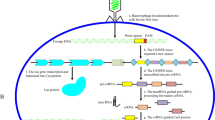
Abstract
This chapter describes the potential use of viral-mediated gene transfer in the central nervous system for genome editing in the context of Huntington’s disease. Here, we provide protocols that cover the design of various genome editing strategies, the cloning of CRISPR/Cas9 elements into lentiviral vectors, and the assessment of cleavage efficiency, as well as potential unwanted effects.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Urnov FD, Rebar EJ, Holmes MC et al (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11:636–646
Im W, Moon J, Kim M (2016) Applications of CRISPR/Cas9 for gene editing in hereditary movement disorders. J Mov Disord 9:136–143
Fagerlund RD, Staals RH, Fineran PC (2015) The Cpf1 CRISPR-Cas protein expands genome-editing tools. Genome Biol 16:251
Ran FA, Cong L, Yan WX et al (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191
Riordan SM, Heruth DP, Zhang LQ, Ye SQ (2015) Application of CRISPR/Cas9 for biomedical discoveries. Cell Biosci 5:33
Wang F, Qi LS (2016) Applications of CRISPR genome engineering in cell biology. Trends Cell Biol 26:875–888
Heyer WD, Ehmsen KT, Liu J (2010) Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44:113–139
Ran FA, Hsu PD, Wright J et al (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Walters BJ, Azam AB, Gillon CJ et al (2015) Advanced in vivo use of CRISPR/Cas9 and anti-sense DNA inhibition for gene manipulation in the brain. Front Genet 6:362
Bates GP, Dorsey R, Gusella JF et al (2015) Huntington disease. Nat Rev Dis Primers 1:15005
Smith A, Zanardi T, Norris D et al (2016) Antisense oligonucleotides enter clinical trials. HD Insights 13:10–15
Boudreau RL, McBride JL, Martins I et al (2009) Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther 17:1053–1063
Drouet V, Perrin V, Hassig R et al (2009) Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol 65:276–285
Wang G, Liu X, Gaertig MA et al (2016) Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc Natl Acad Sci U S A 113:3359–3364
Kay C, Skotte NH, Southwell AL, Hayden MR (2014) Personalized gene silencing therapeutics for Huntington disease. Clin Genet 86:29–36
Lee JM, Gillis T, Mysore JS et al (2012) Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am J Hum Genet 90:434–444
Lee JM, Kim KH, Shin A et al (2015) Sequence-level analysis of the major European Huntington disease haplotype. Am J Hum Genet 97:435–444
Warby SC, Montpetit A, Hayden AR et al (2009) CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet 84:351–366
Warby SC, Visscher H, Collins JA et al (2011) HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur J Hum Genet 19:561–566
Southwell AL, Skotte NH, Bennett CF, Hayden MR (2012) Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol Med 18:634–643. https://doi.org/10.1016/j.molmed.2012.09.001
Aronin N, DiFiglia M (2014) Huntingtin-lowering strategies in Huntington’s disease: antisense oligonucleotides, small RNAs, and gene editing. Mov Disord 29:1455–1461
Drouet V, Ruiz M, Zala D et al (2014) Allele-specific silencing of mutant huntingtin in rodent brain and human stem cells. PLoS One 9:e99341
Skotte NH, Southwell AL, Ostergaard ME et al (2014) Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS One 9:e107434
Shin JW, Kim KH, Chao MJ et al (2016) Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet 25:4566–4576
Monteys AM, Ebanks SA, Keiser MS, Davidson BL (2017) CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol Ther 25:12–23
Liu J, Shui SL (2016) Delivery methods for site-specific nucleases: achieving the full potential of therapeutic gene editing. J Control Release 244:83–97
Wang L, Li F, Dang L et al (2016) In vivo delivery systems for therapeutic genome editing. Int J Mol Sci 17:pii.E626
Delzor A, Dufour N, Deglon N (2014) Lentiviral vectors in Huntington’s disease research and therapy. In: Brambilla R (ed) Viral vector approaches in neurobiology and brain diseases book series, Neuromethods, vol 82. Humana Press, Totowa NJ, pp 193–220
McClure C, Cole KL, Wulff P et al (2011) Production and titering of recombinant adeno-associated viral vectors. J Vis Exp 57:e3348
Merten OW, Hebben M, Bovolenta C (2016) Production of lentiviral vectors. Mol Ther Methods Clin Dev 3:16017
Kim D, Bae S, Park J et al (2015) Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 12:237–243
Slaymaker IM, Gao L, Zetsche B et al (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88
Tsai SQ, Zheng Z, Nguyen NT et al (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33:187–197
Doench JG, Hartenian E, Graham DB et al (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32:1262–1267
Hsu PD, Scott DA, Weinstein JA et al (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832
Fu Y, Sander JD, Reyon D et al (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Tycko J, Myer VE, Hsu PD (2016) Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol Cell 63:355–370
Bolukbasi MF, Gupta A, Oikemus S et al (2015) DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods 12:1150–1156
Mali P, Yang L, Esvelt KM et al (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Zetsche B, Gootenberg JS, Abudayyeh OO et al (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771
Ran FA (2016) Adaptation of CRISPR nucleases for eukaryotic applications. Anal Biochem 532:90–94
Doench JG, Fusi N, Sullender M et al (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191
Sanjana NE, Shalem O, Zhang F (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784
Gray SJ, Foti SB, Schwartz JW et al (2011) Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum Gene Ther 22:1143–1153
Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA (2014) Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res 42:e147
Cong L, Ran FA, Cox D et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Bauer DE, Canver MC, Orkin SH (2015) Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J Vis Exp:e52118. https://doi.org/10.3791/52118
Maddalo D, Manchado E, Concepcion CP et al (2014) In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516:423–427. https://doi.org/10.1038/nature13902
Vouillot L, Thelie A, Pollet N (2015) Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 5:407–415
Brinkman EK, Chen T, Amendola M, van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42:e168
Pinello L, Canver MC, Hoban MD et al (2016) Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol 34:695–697
Smith C, Gore A, Yan W et al (2014) Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15:12–13
Veres A, Gosis BS, Ding Q et al (2014) Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15:27–30
Crosetto N, Mitra A, Silva MJ et al (2013) Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 10:361–365
Dang Y, Jia G, Choi J, Ma H et al (2015) Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol 16:280
Cho SW, Kim S, Kim Y et al (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24:132–141
Kleinstiver BP, Pattanayak V, Prew MS et al (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529:490–495
Shen B, Zhang W, Zhang J et al (2014) Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 11:399–402
Hodgson JG, Agopyan N, Gutekunst CA et al (1999) A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron 23:181–192
Gray M, Shirasaki DI, Cepeda C et al (2008) Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci 28:6182–6195
Southwell AL, Warby SC, Carroll JB et al (2013) A fully humanized transgenic mouse model of Huntington disease. Hum Mol Genet 22:18–34
Southwell AL, Skotte NH, Villanueva EB et al (2017) A novel humanized mouse model of Huntington disease for preclinical development of therapeutics targeting mutant huntingtin alleles. Hum Mol Genet 26:1115–1132
Swiech L, Heidenreich M, Banerjee A et al (2015) In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33:102–106
Waltz E (2016) CRISPR-edited crops free to enter market, skip regulation. Nat Biotechnol 34:582
Waltz E (2016) Gene-edited CRISPR mushroom escapes US regulation. Nature 532:293
Callaway E (2016) BIOTECHNOLOGY Embryo editing gets green light. Nature 530:18
Hayden EC (2016) Tomorrow’s children: what would genome editing really mean for future generations? Nature 530:402–405
Ledford H (2017) Court rules on CRISPR. Nature 542:401
Cyranoski D (2016) CRISPR gene-editing tested in a person for the first time. Nature 539:479
Kleinstiver BP, Prew MS, Tsai SQ et al (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485
Sakuma T, Nishikawa A, Kume S et al (2014) Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep 4:5400
Vad-Nielsen J, Lin L, Bolund L et al (2016) Golden Gate Assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes. Cell Mol Life Sci 73:4315–4325
Jiang W, Zhao X, Gabrieli T et al (2015) Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat Commun 6:8101
Wang JW, Wang A, Li K et al (2015) CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning. Biotechniques 58:161–170
Acknowledgments
We would like to thank Maria Rey for the preparation of the figures.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Vachey, G., Déglon, N. (2018). CRISPR/Cas9-Mediated Genome Editing for Huntington’s Disease. In: Precious, S., Rosser, A., Dunnett, S. (eds) Huntington’s Disease. Methods in Molecular Biology, vol 1780. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7825-0_21
Download citation
DOI: https://doi.org/10.1007/978-1-4939-7825-0_21
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-7824-3
Online ISBN: 978-1-4939-7825-0
eBook Packages: Springer Protocols