Newer gene editing technologies toward HIV gene therapy
- PMID: 24284874
- PMCID: PMC3856413
- DOI: 10.3390/v5112748
Newer gene editing technologies toward HIV gene therapy
Abstract
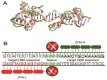
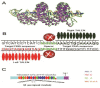
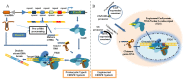
Despite the great success of highly active antiretroviral therapy (HAART) in ameliorating the course of HIV infection, alternative therapeutic approaches are being pursued because of practical problems associated with life-long therapy. The eradication of HIV in the so-called "Berlin patient" who received a bone marrow transplant from a CCR5-negative donor has rekindled interest in genome engineering strategies to achieve the same effect. Precise gene editing within the cells is now a realistic possibility with recent advances in understanding the DNA repair mechanisms, DNA interaction with transcription factors and bacterial defense mechanisms. Within the past few years, four novel technologies have emerged that can be engineered for recognition of specific DNA target sequences to enable site-specific gene editing: Homing Endonuclease, ZFN, TALEN, and CRISPR/Cas9 system. The most recent CRISPR/Cas9 system uses a short stretch of complementary RNA bound to Cas9 nuclease to recognize and cleave target DNA, as opposed to the previous technologies that use DNA binding motifs of either zinc finger proteins or transcription activator-like effector molecules fused to an endonuclease to mediate sequence-specific DNA cleavage. Unlike RNA interference, which requires the continued presence of effector moieties to maintain gene silencing, the newer technologies allow permanent disruption of the targeted gene after a single treatment. Here, we review the applications, limitations and future prospects of novel gene-editing strategies for use as HIV therapy.
Figures
Similar articles
-
Gene Therapy with CRISPR/Cas9 Coming to Age for HIV Cure.AIDS Rev. 2017 Oct-Dec;19(3):167-172. AIDS Rev. 2017. PMID: 29019352
-
The therapeutic application of CRISPR/Cas9 technologies for HIV.Expert Opin Biol Ther. 2015 Jun;15(6):819-30. doi: 10.1517/14712598.2015.1036736. Epub 2015 Apr 12. Expert Opin Biol Ther. 2015. PMID: 25865334 Free PMC article. Review.
-
The therapeutic landscape of HIV-1 via genome editing.AIDS Res Ther. 2017 Jul 14;14(1):32. doi: 10.1186/s12981-017-0157-8. AIDS Res Ther. 2017. PMID: 28705213 Free PMC article. Review.
-
[Application progress of CRISPR/Cas9 genome editing technology in the treatment of HIV-1 infection].Yi Chuan. 2016 Jan;38(1):9-16. doi: 10.16288/j.yczz.15-284. Yi Chuan. 2016. PMID: 26787519 Review. Chinese.
-
Recent advances in RNAi-based strategies for therapy and prevention of HIV-1/AIDS.Adv Drug Deliv Rev. 2016 Aug 1;103:174-186. doi: 10.1016/j.addr.2016.03.005. Epub 2016 Mar 21. Adv Drug Deliv Rev. 2016. PMID: 27013255 Free PMC article. Review.
Cited by
-
Stem cell gene therapy for HIV: strategies to inhibit viral entry and replication.Curr HIV/AIDS Rep. 2015 Mar;12(1):79-87. doi: 10.1007/s11904-014-0242-8. Curr HIV/AIDS Rep. 2015. PMID: 25578054 Review.
-
The CRISPR/Cas9 genome editing methodology as a weapon against human viruses.Discov Med. 2015 Apr;19(105):255-62. Discov Med. 2015. PMID: 25977188 Free PMC article. Review.
-
Inhibition of HIV-1 Viral Infection by an Engineered CRISPR Csy4 RNA Endoribonuclease.PLoS One. 2015 Oct 23;10(10):e0141335. doi: 10.1371/journal.pone.0141335. eCollection 2015. PLoS One. 2015. PMID: 26495836 Free PMC article.
-
TALEN gene editing takes aim on HIV.Hum Genet. 2016 Sep;135(9):1059-70. doi: 10.1007/s00439-016-1678-2. Epub 2016 May 12. Hum Genet. 2016. PMID: 27170155 Free PMC article. Review.
-
Engineering T Cells to Functionally Cure HIV-1 Infection.Mol Ther. 2015 Jul;23(7):1149-1159. doi: 10.1038/mt.2015.70. Epub 2015 Apr 21. Mol Ther. 2015. PMID: 25896251 Free PMC article. Review.
References
-
- Zaccarelli M., Tozzi V., Lorenzini P., Trotta M.P., Forbici F., Visco-Comandini U., Gori C., Narciso P., Perno C.F., Antinori A., et al. Multiple drug class-wide resistance associated with poorer survival after treatment failure in a cohort of HIV-infected patients. AIDS. 2005;19:1081–1089. doi: 10.1097/01.aids.0000174455.01369.ad. - DOI - PubMed
-
- Finzi D., Blankson J., Siliciano J.D., Margolick J.B., Chadwick K., Pierson T., Smith K., Lisziewicz J., Lori F., Flexner C., et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999;5:512–517. doi: 10.1038/8394. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

