Distinct methylation of the interferon gamma (IFN-gamma) and interleukin 3 (IL-3) genes in newly activated primary CD8+ T lymphocytes: regional IFN-gamma promoter demethylation and mRNA expression are heritable in CD44(high)CD8+ T cells
- PMID: 9653088
- PMCID: PMC2525536
- DOI: 10.1084/jem.188.1.103
Distinct methylation of the interferon gamma (IFN-gamma) and interleukin 3 (IL-3) genes in newly activated primary CD8+ T lymphocytes: regional IFN-gamma promoter demethylation and mRNA expression are heritable in CD44(high)CD8+ T cells
Abstract
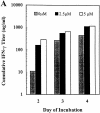
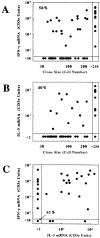
Differential genomic DNA methylation has the potential to influence the development of T cell cytokine production profiles. Therefore, we have conducted a clonal analysis of interferon (IFN)-gamma and interleukin (IL)-3 gene methylation and messenger (m)RNA expression in primary CD8+ T cells during the early stages of activation, growth, and cytokine expression. Despite similar distributions and densities of CpG methylation sites, the IFN-gamma and IL-3 promoters exhibited differential demethylation in the same T cell clone, and heterogeneity between clones. Methylation patterns and mRNA levels were correlated for both genes, but demethylation of the IFN-gamma promoter was widespread across >300 basepairs in clones expressing high levels of IFN-gamma mRNA, whereas demethylation of the IL-3 promoter was confined to specific CpG sites in the same clones. Conversely, the majority of clones expressing low or undetectable levels of IFN-gamma mRNA exhibited symmetrical methylation of four to six of the IFN-gamma promoter CpG sites. Genomic DNA methylation also has the potential to influence the maintenance or stability of T cell cytokine production profiles. Therefore, we also tested the heritability of IFN-gamma gene methylation and mRNA expression in families of clones derived from resting CD44(low)CD8+ T cells or from previously activated CD44(high)CD8+ T cells. The patterns of IFN-gamma gene demethylation and mRNA expression were faithfully inherited in all clones derived from CD44(high) cells, but variable in clones derived from CD44(low) cells. Overall, these findings suggest that differential genomic DNA methylation, including differences among cytokine genes, among individual T cells, and among T cells with different activation histories, is an important feature of cytokine gene expression in primary T cells.
Figures







Similar articles
-
Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells.J Immunol. 2006 Apr 1;176(7):4083-93. doi: 10.4049/jimmunol.176.7.4083. J Immunol. 2006. PMID: 16547244
-
Differential interferon-γ production by naive and memory-like CD8 T cells.J Leukoc Biol. 2020 Oct;108(4):1329-1337. doi: 10.1002/JLB.2AB0420-646R. Epub 2020 May 18. J Leukoc Biol. 2020. PMID: 32421902
-
Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO- T cells.J Immunol. 2002 Mar 15;168(6):2820-7. doi: 10.4049/jimmunol.168.6.2820. J Immunol. 2002. PMID: 11884451
-
Epigenetic control of interferon-gamma expression in CD8 T cells.J Immunol Res. 2015;2015:849573. doi: 10.1155/2015/849573. Epub 2015 Apr 20. J Immunol Res. 2015. PMID: 25973438 Free PMC article. Review.
-
Molecular regulation of cytokine gene expression: interferon-gamma as a model system.Prog Nucleic Acid Res Mol Biol. 1997;56:109-27. doi: 10.1016/s0079-6603(08)61004-1. Prog Nucleic Acid Res Mol Biol. 1997. PMID: 9187053 Review.
Cited by
-
STAT4 and T-bet are required for the plasticity of IFN-γ expression across Th2 ontogeny and influence changes in Ifng promoter DNA methylation.J Immunol. 2013 Jul 15;191(2):678-87. doi: 10.4049/jimmunol.1203360. Epub 2013 Jun 12. J Immunol. 2013. PMID: 23761633 Free PMC article.
-
The cytokine response to physical activity and training.Sports Med. 2001 Feb;31(2):115-44. doi: 10.2165/00007256-200131020-00004. Sports Med. 2001. PMID: 11227979 Free PMC article. Review.
-
DNA methylation maintains allele-specific KIR gene expression in human natural killer cells.J Exp Med. 2003 Jan 20;197(2):245-55. doi: 10.1084/jem.20021127. J Exp Med. 2003. PMID: 12538663 Free PMC article.
-
Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse.J Clin Invest. 2003 Feb;111(4):539-52. doi: 10.1172/JCI16153. J Clin Invest. 2003. PMID: 12588892 Free PMC article.
-
Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence.EMBO J. 2011 Jan 19;30(2):263-76. doi: 10.1038/emboj.2010.314. Epub 2010 Dec 17. EMBO J. 2011. PMID: 21169989 Free PMC article.
References
-
- Kelso A. Th1 and Th2 subsets: paradigms lost? . Immunol Today. 1995;16:372–379. - PubMed
-
- Kelso A, Groves P, Troutt AB, Francis K. Evidence for the stochastic acquisition of cytokine profile by CD4+ T cells activated in a T helper type 2-like response in vivo. . Eur J Immunol. 1995;25:1168–1175. - PubMed
-
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–791. - PubMed
-
- Ernst P, Smale ST. Combinatorial regulation of transcription I: general aspects of transcriptional control. Immunity. 1995;2:311–319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

