T cell egress via lymphatic vessels is tuned by antigen encounter and limits tumor control
- PMID: 36849745
- PMCID: PMC10998279
- DOI: 10.1038/s41590-023-01443-y
T cell egress via lymphatic vessels is tuned by antigen encounter and limits tumor control
Erratum in
-
Author Correction: T cell egress via lymphatic vessels is tuned by antigen encounter and limits tumor control.Nat Immunol. 2023 Apr;24(4):729. doi: 10.1038/s41590-023-01491-4. Nat Immunol. 2023. PMID: 36932125 No abstract available.
Abstract
Antigen-specific CD8+ T cell accumulation in tumors is a prerequisite for effective immunotherapy, and yet the mechanisms of lymphocyte transit are not well defined. Here we show that tumor-associated lymphatic vessels control T cell exit from tumors via the chemokine CXCL12, and intratumoral antigen encounter tunes CXCR4 expression by effector CD8+ T cells. Only high-affinity antigen downregulates CXCR4 and upregulates the CXCL12 decoy receptor, ACKR3, thereby reducing CXCL12 sensitivity and promoting T cell retention. A diverse repertoire of functional tumor-specific CD8+ T cells, therefore, exit the tumor, which limits the pool of CD8+ T cells available to exert tumor control. CXCR4 inhibition or loss of lymphatic-specific CXCL12 boosts T cell retention and enhances tumor control. These data indicate that strategies to limit T cell egress might be an approach to boost the quantity and quality of intratumoral T cells and thereby response to immunotherapy.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Figures
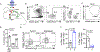
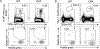


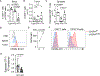










Similar articles
-
CXCR4 is dispensable for T cell egress from chronically inflamed skin via the afferent lymph.PLoS One. 2014 Apr 21;9(4):e95626. doi: 10.1371/journal.pone.0095626. eCollection 2014. PLoS One. 2014. PMID: 24752354 Free PMC article.
-
Murine B16 melanomas expressing high levels of the chemokine stromal-derived factor-1/CXCL12 induce tumor-specific T cell chemorepulsion and escape from immune control.J Immunol. 2006 Mar 1;176(5):2902-14. doi: 10.4049/jimmunol.176.5.2902. J Immunol. 2006. PMID: 16493048
-
CXCR4/ACKR3/CXCL12 axis in the lymphatic metastasis of vulvar squamous cell carcinoma.J Clin Pathol. 2022 May;75(5):324-332. doi: 10.1136/jclinpath-2020-206917. Epub 2021 Mar 10. J Clin Pathol. 2022. PMID: 33692092 Free PMC article.
-
Potential Role of CXCR4 Targeting in the Context of Radiotherapy and Immunotherapy of Cancer.Front Immunol. 2018 Dec 21;9:3018. doi: 10.3389/fimmu.2018.03018. eCollection 2018. Front Immunol. 2018. PMID: 30622535 Free PMC article. Review.
-
CXCR4/CXCL12 Activities in the Tumor Microenvironment and Implications for Tumor Immunotherapy.Cancers (Basel). 2022 May 6;14(9):2314. doi: 10.3390/cancers14092314. Cancers (Basel). 2022. PMID: 35565443 Free PMC article. Review.
Cited by
-
The tumor-draining lymph node as a reservoir for systemic immune surveillance.Trends Cancer. 2024 Jan;10(1):28-37. doi: 10.1016/j.trecan.2023.09.006. Epub 2023 Oct 18. Trends Cancer. 2024. PMID: 37863720 Free PMC article. Review.
-
T Cell Surveillance during Cutaneous Viral Infections.Viruses. 2024 Apr 26;16(5):679. doi: 10.3390/v16050679. Viruses. 2024. PMID: 38793562 Free PMC article. Review.
-
Pan-cancer integrative analyses dissect the remodeling of endothelial cells in human cancers.Natl Sci Rev. 2024 Jul 11;11(9):nwae231. doi: 10.1093/nsr/nwae231. eCollection 2024 Sep. Natl Sci Rev. 2024. PMID: 39345334 Free PMC article.
-
Dendritic cells as shepherds of T cell immunity in cancer.Immunity. 2023 Oct 10;56(10):2218-2230. doi: 10.1016/j.immuni.2023.08.014. Epub 2023 Sep 13. Immunity. 2023. PMID: 37708889 Free PMC article. Review.
-
A seven-immune-genes risk model predicts the survival and suitable treatments for patients with skin cutaneous melanoma.Heliyon. 2023 Sep 19;9(9):e20234. doi: 10.1016/j.heliyon.2023.e20234. eCollection 2023 Sep. Heliyon. 2023. PMID: 37809963 Free PMC article.
References
Methods-only References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

