Senescence atlas reveals an aged-like inflamed niche that blunts muscle regeneration
- PMID: 36544018
- PMCID: PMC9812788
- DOI: 10.1038/s41586-022-05535-x
Senescence atlas reveals an aged-like inflamed niche that blunts muscle regeneration
Erratum in
-
Author Correction: Senescence atlas reveals an aged-like inflamed niche that blunts muscle regeneration.Nature. 2023 Feb;614(7949):E45. doi: 10.1038/s41586-023-05765-7. Nature. 2023. PMID: 36747036 Free PMC article. No abstract available.
Abstract
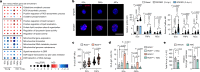
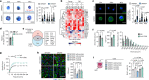
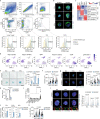
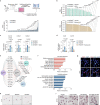
Tissue regeneration requires coordination between resident stem cells and local niche cells1,2. Here we identify that senescent cells are integral components of the skeletal muscle regenerative niche that repress regeneration at all stages of life. The technical limitation of senescent-cell scarcity3 was overcome by combining single-cell transcriptomics and a senescent-cell enrichment sorting protocol. We identified and isolated different senescent cell types from damaged muscles of young and old mice. Deeper transcriptome, chromatin and pathway analyses revealed conservation of cell identity traits as well as two universal senescence hallmarks (inflammation and fibrosis) across cell type, regeneration time and ageing. Senescent cells create an aged-like inflamed niche that mirrors inflammation associated with ageing (inflammageing4) and arrests stem cell proliferation and regeneration. Reducing the burden of senescent cells, or reducing their inflammatory secretome through CD36 neutralization, accelerates regeneration in young and old mice. By contrast, transplantation of senescent cells delays regeneration. Our results provide a technique for isolating in vivo senescent cells, define a senescence blueprint for muscle, and uncover unproductive functional interactions between senescent cells and stem cells in regenerative niches that can be overcome. As senescent cells also accumulate in human muscles, our findings open potential paths for improving muscle repair throughout life.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures














Comment in
-
Senescent cells damage the body throughout life.Nature. 2023 Jan;613(7942):30-31. doi: 10.1038/d41586-022-04430-9. Nature. 2023. PMID: 36544002 No abstract available.
Similar articles
-
Restoration of hippocampal neural precursor function by ablation of senescent cells in the aging stem cell niche.Stem Cell Reports. 2022 Feb 8;17(2):259-275. doi: 10.1016/j.stemcr.2021.12.010. Epub 2022 Jan 20. Stem Cell Reports. 2022. PMID: 35063124 Free PMC article.
-
Muscle injury induces a transient senescence-like state that is required for myofiber growth during muscle regeneration.FASEB J. 2022 Nov;36(11):e22587. doi: 10.1096/fj.202200289RR. FASEB J. 2022. PMID: 36190443
-
Geroconversion of aged muscle stem cells under regenerative pressure.Cell Cycle. 2014;13(20):3183-90. doi: 10.4161/15384101.2014.965072. Cell Cycle. 2014. PMID: 25485497 Free PMC article.
-
Muscle stem cell niche dynamics during muscle homeostasis and regeneration.Curr Top Dev Biol. 2024;158:151-177. doi: 10.1016/bs.ctdb.2024.02.008. Epub 2024 Mar 23. Curr Top Dev Biol. 2024. PMID: 38670704 Review.
-
The right time for senescence.Elife. 2021 Nov 10;10:e72449. doi: 10.7554/eLife.72449. Elife. 2021. PMID: 34756162 Free PMC article. Review.
Cited by
-
Calcium's Role and Signaling in Aging Muscle, Cellular Senescence, and Mineral Interactions.Int J Mol Sci. 2023 Dec 1;24(23):17034. doi: 10.3390/ijms242317034. Int J Mol Sci. 2023. PMID: 38069357 Free PMC article. Review.
-
Aging research comes of age.Nat Methods. 2024 Jan;21(1):11-15. doi: 10.1038/s41592-023-02140-2. Nat Methods. 2024. PMID: 38167657 No abstract available.
-
Inhibition of p53-MDM2 binding reduces senescent cell abundance and improves the adaptive responses of skeletal muscle from aged mice.Geroscience. 2024 Apr;46(2):2153-2176. doi: 10.1007/s11357-023-00976-2. Epub 2023 Oct 24. Geroscience. 2024. PMID: 37872294 Free PMC article.
-
Realveolarization with inhalable mucus-penetrating lipid nanoparticles for the treatment of pulmonary fibrosis in mice.Sci Adv. 2024 Jun 14;10(24):eado4791. doi: 10.1126/sciadv.ado4791. Epub 2024 Jun 12. Sci Adv. 2024. PMID: 38865465 Free PMC article.
-
The life and times of cellular senescence in skeletal muscle: friend or foe for homeostasis and adaptation?Am J Physiol Cell Physiol. 2023 Jul 1;325(1):C324-C331. doi: 10.1152/ajpcell.00553.2022. Epub 2023 Jun 19. Am J Physiol Cell Physiol. 2023. PMID: 37335024 Free PMC article. Review.
References
-
- Sousa-Victor P, Garcia-Prat L, Munoz-Canoves P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 2022;23:204–226. - PubMed
-
- Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A. 2014;69:S4–S9. - PubMed
-
- Sousa-Victor P, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

