Identification of miRNA-mRNA Pairs in the Alzheimer's Disease Expression Profile and Explore the Effect of miR-26a-5p/PTGS2 on Amyloid-β Induced Neurotoxicity in Alzheimer's Disease Cell Model
- PMID: 35783137
- PMCID: PMC9249435
- DOI: 10.3389/fnagi.2022.909222
Identification of miRNA-mRNA Pairs in the Alzheimer's Disease Expression Profile and Explore the Effect of miR-26a-5p/PTGS2 on Amyloid-β Induced Neurotoxicity in Alzheimer's Disease Cell Model
Abstract
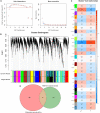
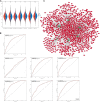
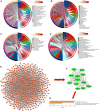
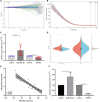
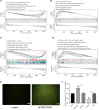
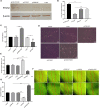
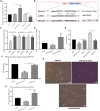
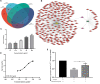
Alzheimer's disease (AD) is a progressive neurodegenerative disease and the most common type of dementia. MicroRNAs (miRNAs) have been extensively studied in many diseases, including AD. To identify the AD-specific differentially expressed miRNAs and mRNAs, we used bioinformatics analysis to study candidate miRNA-mRNA pairs involved in the pathogenesis of AD. These miRNA-mRNAs may serve as promising biomarkers for early diagnosis or targeted therapy of AD patients. In this study, based on the AD mRNA and miRNA expression profile data in Gene Expression Omnibus (GEO), through differential expression analysis, functional annotation and enrichment analysis, weighted gene co-expression network analysis, miRNA-mRNA regulatory network, protein-protein interaction network, receiver operator characteristic and Least absolute shrinkage and selection operator (LASSO) regression and other analysis, we screened the key miRNA-mRNA in the progress of AD: miR-26a-5p/PTGS2. Dual-luciferase and qPCR experiments confirmed that PTGS2 is a direct target gene of miR-26a-5p. The expression of miR-26a-5p in the peripheral blood of AD patients and AD model cells (SH-SY5Y cells treated with Aβ25-35) was up-regulated, and the expression of PTGS2 was down-regulated. Functional gain -loss experiments confirmed that PTGS2 protects AD model cells from damage by inhibiting proliferation and migration. However, the expression of miR-26a-5p promotes the proliferation of AD model cells. It is further found that PTGS2 is involved in the regulation of miR-26a-5p and can reverse the effect of miR-26a-5p on the proliferation of AD model cells. In addition, through network pharmacology, qPCR and CCK-8, we found that baicalein may affect the progression of AD by regulating the expression of PTGS2. Therefore, PTGS2 can be used as a target for AD research, and miR-26a-5p/PTGS2 can be used as an axis of action to study the pathogenesis of AD.
Keywords: Alzheimer’s disease; baicalein; miR-26a-5p/PTGS2; miRNA–mRNA; neurotoxicity.
Copyright © 2022 Xie, Pei, Shan, Xiao, Zhou, Huang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Genome-Wide Profiling of miRNA and mRNA Expression in Alzheimer's Disease.Med Sci Monit. 2017 Jun 4;23:2721-2731. doi: 10.12659/msm.905064. Med Sci Monit. 2017. PMID: 28578378 Free PMC article.
-
EPC-exosomal miR-26a-5p improves airway remodeling in COPD by inhibiting ferroptosis of bronchial epithelial cells via PTGS2/PGE2 signaling pathway.Sci Rep. 2023 Apr 14;13(1):6126. doi: 10.1038/s41598-023-33151-w. Sci Rep. 2023. PMID: 37059741 Free PMC article.
-
Overexpression of miR-26a-5p Suppresses Tau Phosphorylation and Aβ Accumulation in the Alzheimer's Disease Mice by Targeting DYRK1A.Curr Neurovasc Res. 2020;17(3):241-248. doi: 10.2174/1567202617666200414142637. Curr Neurovasc Res. 2020. PMID: 32286945
-
The Eminent Role of microRNAs in the Pathogenesis of Alzheimer's Disease.Front Aging Neurosci. 2021 Mar 15;13:641080. doi: 10.3389/fnagi.2021.641080. eCollection 2021. Front Aging Neurosci. 2021. PMID: 33790780 Free PMC article. Review.
-
Exploiting machine learning models to identify novel Alzheimer's disease biomarkers and potential targets.Sci Rep. 2023 Mar 27;13(1):4979. doi: 10.1038/s41598-023-30904-5. Sci Rep. 2023. PMID: 36973386 Free PMC article. Review.
Cited by
-
Exploring the pathogenesis linking traumatic brain injury and epilepsy via bioinformatic analyses.Front Aging Neurosci. 2022 Nov 10;14:1047908. doi: 10.3389/fnagi.2022.1047908. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36438009 Free PMC article.
-
Identifying potential drug-target interactions based on ensemble deep learning.Front Aging Neurosci. 2023 Jun 15;15:1176400. doi: 10.3389/fnagi.2023.1176400. eCollection 2023. Front Aging Neurosci. 2023. PMID: 37396659 Free PMC article.
-
Bibliometric analysis of research on Alzheimer's disease and non-coding RNAs: Opportunities and challenges.Front Aging Neurosci. 2022 Oct 18;14:1037068. doi: 10.3389/fnagi.2022.1037068. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36329875 Free PMC article.
-
Expression and Regulatory Mechanisms of MicroRNA in Cholesteatoma: A Systematic Review.Int J Mol Sci. 2023 Jul 31;24(15):12277. doi: 10.3390/ijms241512277. Int J Mol Sci. 2023. PMID: 37569652 Free PMC article. Review.
-
MicroRNAs-Based Theranostics against Anesthetic-Induced Neurotoxicity.Pharmaceutics. 2023 Jun 27;15(7):1833. doi: 10.3390/pharmaceutics15071833. Pharmaceutics. 2023. PMID: 37514018 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials

