Therapeutic Targeting of EZH2 and BET BRD4 in Pediatric Rhabdoid Tumors
- PMID: 35247919
- PMCID: PMC9081147
- DOI: 10.1158/1535-7163.MCT-21-0646
Therapeutic Targeting of EZH2 and BET BRD4 in Pediatric Rhabdoid Tumors
Abstract
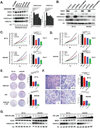
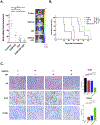
Aberrant activity of the H3K27 modifiers EZH2 and BRD4 is an important oncogenic driver for atypical teratoid/rhabdoid tumor (AT/RT), and each is potentially a possible therapeutic target for treating AT/RT. We, therefore, determined whether targeting distinct histone modifier activities was an effective approach for treating AT/RT. The effects of EZH2 and BRD4 inhibition on histone modification, cell proliferation, and cell invasion were analyzed by immunoblotting, MTS assay, colony formation assay, and cell invasion assay. RNA- and chromatin immunoprecipitation-sequencing were used to determine transcriptional and epigenetic changes in AT/RT cells treated with EZH2 and BRD4 inhibitors. We treated mice bearing human AT/RT xenografts with EZH2 and BRD4 inhibitors. Intracranial tumor growth was monitored by bioluminescence imaging, and the therapeutic response was evaluated by animal survival. AT/RT cells showed elevated levels of H3K27 trimethylation (H3K27me3) and H3K27 acetylation (H3K27ac), with expression of EZH2 and BRD4, and lack of SMARCB1 proteins. Targeted inhibition of EZH2 and BRD4 activities reduced cell proliferation and invasiveness of AT/RT in association with decreasing H3K27me3 and H3K27ac. Differential genomic occupancy of H3K27me3 and H3K27ac regulated specific gene expression in response to EZH2 and BRD4 inhibitions. A combination of EZH2 and BRD4 inhibition increased the therapeutic benefit in vitro and in vivo, outperforming either monotherapy. Overall, histones H3K27me3 and H3K27ac were elevated in AT/RT cells and distributed in distinct chromatin regions to regulate specific gene expression and to promote AT/RT growth. Targeting EZH2 and BRD4 activity is, therefore, a potential combination therapy for AT/RT.
©2022 American Association for Cancer Research.
Conflict of interest statement
CONFLICT OF INTEREST
The authors have declared that no conflict of interest exists.
Figures


Similar articles
-
EZH2 Cooperates with BRD4-NUT to Drive NUT Carcinoma Growth by Silencing Key Tumor Suppressor Genes.Cancer Res. 2023 Dec 1;83(23):3956-3973. doi: 10.1158/0008-5472.CAN-23-1475. Cancer Res. 2023. PMID: 37747726 Free PMC article.
-
Design and Synthesis of Dual EZH2/BRD4 Inhibitors to Target Solid Tumors.J Med Chem. 2022 May 12;65(9):6573-6592. doi: 10.1021/acs.jmedchem.1c01876. Epub 2022 May 2. J Med Chem. 2022. PMID: 35500243
-
Targeting Epigenetic Crosstalk as a Therapeutic Strategy for EZH2-Aberrant Solid Tumors.Cell. 2018 Sep 20;175(1):186-199.e19. doi: 10.1016/j.cell.2018.08.058. Epub 2018 Sep 13. Cell. 2018. PMID: 30220457
-
Recent advances in EZH2-based dual inhibitors in the treatment of cancers.Eur J Med Chem. 2023 Aug 5;256:115461. doi: 10.1016/j.ejmech.2023.115461. Epub 2023 May 5. Eur J Med Chem. 2023. PMID: 37156182 Review.
-
EZH2-Targeted Therapies in Cancer: Hype or a Reality.Cancer Res. 2020 Dec 15;80(24):5449-5458. doi: 10.1158/0008-5472.CAN-20-2147. Epub 2020 Sep 25. Cancer Res. 2020. PMID: 32978169 Free PMC article. Review.
Cited by
-
Development and epigenetic regulation of Atypical teratoid/rhabdoid tumors in the context of cell-of-origin and halted cell differentiation.Neurooncol Adv. 2024 Sep 23;6(1):vdae162. doi: 10.1093/noajnl/vdae162. eCollection 2024 Jan-Dec. Neurooncol Adv. 2024. PMID: 39465218 Free PMC article. Review.
-
Epigenetic therapies targeting histone lysine methylation: complex mechanisms and clinical challenges.J Clin Invest. 2024 Oct 15;134(20):e183391. doi: 10.1172/JCI183391. J Clin Invest. 2024. PMID: 39403928 Free PMC article. Review.
-
Targeted Epigenetic Interventions in Cancer with an Emphasis on Pediatric Malignancies.Biomolecules. 2022 Dec 28;13(1):61. doi: 10.3390/biom13010061. Biomolecules. 2022. PMID: 36671446 Free PMC article. Review.
-
Current advances in immunotherapy for atypical teratoid rhabdoid tumor (ATRT).Neurooncol Pract. 2023 Jan 28;10(4):322-334. doi: 10.1093/nop/npad005. eCollection 2023 Aug. Neurooncol Pract. 2023. PMID: 37457224 Free PMC article. Review.
-
Clinicopathological and molecular characterization of a case classified by DNA‑methylation profiling as "CNS embryonal tumor with BRD4-LEUTX fusion".Acta Neuropathol Commun. 2023 Mar 18;11(1):46. doi: 10.1186/s40478-023-01549-2. Acta Neuropathol Commun. 2023. PMID: 36934287 Free PMC article.
References
-
- Nemes K, Frühwald MC. Emerging therapeutic targets for the treatment of malignant rhabdoid tumors. Expert Opin Ther Targets. 2018;22(4):365–379. - PubMed
-
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Aurias A, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. - PubMed
-
- Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

