Altered succinylation of mitochondrial proteins, APP and tau in Alzheimer's disease
- PMID: 35013160
- PMCID: PMC8748865
- DOI: 10.1038/s41467-021-27572-2
Altered succinylation of mitochondrial proteins, APP and tau in Alzheimer's disease
Abstract
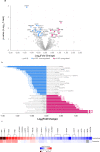
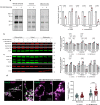
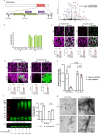
Abnormalities in brain glucose metabolism and accumulation of abnormal protein deposits called plaques and tangles are neuropathological hallmarks of Alzheimer's disease (AD), but their relationship to disease pathogenesis and to each other remains unclear. Here we show that succinylation, a metabolism-associated post-translational protein modification (PTM), provides a potential link between abnormal metabolism and AD pathology. We quantified the lysine succinylomes and proteomes from brains of individuals with AD, and healthy controls. In AD, succinylation of multiple mitochondrial proteins declined, and succinylation of small number of cytosolic proteins increased. The largest increases occurred at critical sites of amyloid precursor protein (APP) and microtubule-associated tau. We show that in vitro, succinylation of APP disrupted its normal proteolytic processing thereby promoting Aβ accumulation and plaque formation and that succinylation of tau promoted its aggregation to tangles and impaired microtubule assembly. In transgenic mouse models of AD, elevated succinylation associated with soluble and insoluble APP derivatives and tau. These findings indicate that a metabolism-linked PTM may be associated with AD.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
[Alzheimer disease: cellular and molecular aspects].Bull Mem Acad R Med Belg. 2005;160(10-12):445-9; discussion 450-1. Bull Mem Acad R Med Belg. 2005. PMID: 16768248 French.
-
Aβ-induced acceleration of Alzheimer-related τ-pathology spreading and its association with prion protein.Acta Neuropathol. 2019 Dec;138(6):913-941. doi: 10.1007/s00401-019-02053-5. Epub 2019 Aug 14. Acta Neuropathol. 2019. PMID: 31414210
-
Abnormal interaction of VDAC1 with amyloid beta and phosphorylated tau causes mitochondrial dysfunction in Alzheimer's disease.Hum Mol Genet. 2012 Dec 1;21(23):5131-46. doi: 10.1093/hmg/dds360. Epub 2012 Aug 27. Hum Mol Genet. 2012. PMID: 22926141 Free PMC article.
-
Exploring the Role of Aggregated Proteomes in the Pathogenesis of Alzheimer's Disease.Curr Protein Pept Sci. 2020;21(12):1164-1173. doi: 10.2174/1389203721666200921152246. Curr Protein Pept Sci. 2020. PMID: 32957903 Review.
-
Proteolytic processing of Alzheimer's β-amyloid precursor protein.J Neurochem. 2012 Jan;120 Suppl 1(Suppl 1):9-21. doi: 10.1111/j.1471-4159.2011.07519.x. Epub 2011 Nov 28. J Neurochem. 2012. PMID: 22122372 Free PMC article. Review.
Cited by
-
Protein modification in neurodegenerative diseases.MedComm (2020). 2024 Aug 4;5(8):e674. doi: 10.1002/mco2.674. eCollection 2024 Aug. MedComm (2020). 2024. PMID: 39105197 Free PMC article. Review.
-
Protein succinylation: regulating metabolism and beyond.Front Nutr. 2024 Feb 6;11:1336057. doi: 10.3389/fnut.2024.1336057. eCollection 2024. Front Nutr. 2024. PMID: 38379549 Free PMC article. Review.
-
Novel histone post-translational modifications in Alzheimer's disease: current advances and implications.Clin Epigenetics. 2024 Mar 9;16(1):39. doi: 10.1186/s13148-024-01650-w. Clin Epigenetics. 2024. PMID: 38461320 Free PMC article. Review.
-
Understanding emerging bioactive metabolites with putative roles in cancer biology.Front Oncol. 2022 Sep 29;12:1014748. doi: 10.3389/fonc.2022.1014748. eCollection 2022. Front Oncol. 2022. PMID: 36249070 Free PMC article. Review.
-
Roles of protein post-translational modifications in glucose and lipid metabolism: mechanisms and perspectives.Mol Med. 2023 Jul 6;29(1):93. doi: 10.1186/s10020-023-00684-9. Mol Med. 2023. PMID: 37415097 Free PMC article. Review.
References
-
- Tjernberg LO, et al. Arrest of -amyloid fibril formation by a pentapeptide ligand. J. Biol. Chem. 1996;271:8545–8548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RF1 AG058469/AG/NIA NIH HHS/United States
- R01 AG022547/AG/NIA NIH HHS/United States
- RF1 AG059319/AG/NIA NIH HHS/United States
- R01 EY026576/EY/NEI NIH HHS/United States
- HHSN271201300031C/DA/NIDA NIH HHS/United States
- S10 OD016320/OD/NIH HHS/United States
- P01 AG014930/AG/NIA NIH HHS/United States
- R01 AG018877/AG/NIA NIH HHS/United States
- R56 AG058469/AG/NIA NIH HHS/United States
- F31 AG069416/AG/NIA NIH HHS/United States
- RF1 AG066493/AG/NIA NIH HHS/United States
- R37 AG019391/AG/NIA NIH HHS/United States
- R01 EY029796/EY/NEI NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- S10 OD017992/OD/NIH HHS/United States
- P30 AG066514/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

