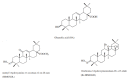
Oleanolic Acid's Semisynthetic Derivatives HIMOXOL and Br-HIMOLID Show Proautophagic Potential and Inhibit Migration of HER2-Positive Breast Cancer Cells In Vitro
- PMID: 34681931
- PMCID: PMC8538366
- DOI: 10.3390/ijms222011273
Oleanolic Acid's Semisynthetic Derivatives HIMOXOL and Br-HIMOLID Show Proautophagic Potential and Inhibit Migration of HER2-Positive Breast Cancer Cells In Vitro
Abstract
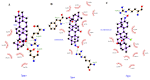
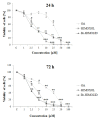
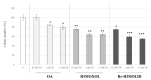
Approximately 20-30% of the diagnosed breast cancers overexpress the human epidermal growth factor receptor 2 (HER2). This type of cancer is associated with a more aggressive phenotype; thus, there is a need for the discovery of new compounds that would improve the survival in HER2-positive breast cancer patients. It seems that one of the most promising therapeutic cancer strategies could be based on the biological activity of pentacyclic triterpenes' derivatives and the best-known representative of this group, oleanolic acid (OA). The biological activity of oleanolic acid and its two semisynthetic derivatives, methyl 3-hydroxyimino-11-oxoolean-12-en-28-oate (HIMOXOL) and 12α-bromo-3-hydroxyimonoolean-28→13-olide (Br-HIMOLID), was assessed in SK-BR-3 breast cancer cells (HER2-positive). Viability tests, cell cycle assessment, evaluation of apoptosis, autophagy, and adhesion/migration processes were performed using MTT, clonogenic, cytofluorometry, Western blot, and qPCR. Both derivatives revealed higher cytotoxicity in studied breast cancer cells than the maternal compound, OA. They also decreased cell viability, induced autophagy, and (when applied in sub-cytotoxic concentrations) decreased the migration of SK-BR-3 cells.This study is the first to report the cytostatic, proautophagic (mTOR/LC3/SQSTM/BECN1 pathway), and anti-migratory (integrin β1/FAK/paxillin pathway) activities of HIMOXOL and Br-HIMOLID in HER2-positive breast cancer cells.
Keywords: Br-HIMOLID; HER2; HIMOXOL; autophagy; breast cancer; migration; oleanolic acid; oleanolic acid derivatives.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Biological Activity of Oleanolic Acid Derivatives HIMOXOL and Br-HIMOLID in Breast Cancer Cells Is Mediated by ER and EGFR.Int J Mol Sci. 2023 Mar 7;24(6):5099. doi: 10.3390/ijms24065099. Int J Mol Sci. 2023. PMID: 36982173 Free PMC article.
-
Methyl 3-hydroxyimino-11-oxoolean-12-en-28-oate (HIMOXOL), a synthetic oleanolic acid derivative, induces both apoptosis and autophagy in MDA-MB-231 breast cancer cells.Chem Biol Interact. 2014 Feb 5;208:47-57. doi: 10.1016/j.cbi.2013.11.009. Epub 2013 Nov 27. Chem Biol Interact. 2014. PMID: 24291674
-
Semisynthetic oleanane triterpenoids inhibit migration and invasion of human breast cancer cells through downregulated expression of the ITGB1/PTK2/PXN pathway.Chem Biol Interact. 2017 Apr 25;268:136-147. doi: 10.1016/j.cbi.2017.03.008. Epub 2017 Mar 18. Chem Biol Interact. 2017. PMID: 28322779
-
Oleanolic Acid and Its Derivatives: Biological Activities and Therapeutic Potential in Chronic Diseases.Molecules. 2017 Nov 13;22(11):1915. doi: 10.3390/molecules22111915. Molecules. 2017. PMID: 29137205 Free PMC article. Review.
-
Oleanane-, ursane-, and quinone methide friedelane-type triterpenoid derivatives: Recent advances in cancer treatment.Eur J Med Chem. 2017 Dec 15;142:95-130. doi: 10.1016/j.ejmech.2017.07.013. Epub 2017 Jul 14. Eur J Med Chem. 2017. PMID: 28754470 Review.
Cited by
-
Doxorubicin and Cisplatin Modulate miR-21, miR-106, miR-126, miR-155 and miR-199 Levels in MCF7, MDA-MB-231 and SK-BR-3 Cells That Makes Them Potential Elements of the DNA-Damaging Drug Treatment Response Monitoring in Breast Cancer Cells-A Preliminary Study.Genes (Basel). 2023 Mar 12;14(3):702. doi: 10.3390/genes14030702. Genes (Basel). 2023. PMID: 36980974 Free PMC article.
-
The Effect of Oleanolic Acid and Its Four New Semisynthetic Derivatives on Human MeWo and A375 Melanoma Cell Lines.Pharmaceuticals (Basel). 2023 May 14;16(5):746. doi: 10.3390/ph16050746. Pharmaceuticals (Basel). 2023. PMID: 37242529 Free PMC article.
-
NOTCH Signaling in Osteosarcoma.Curr Issues Mol Biol. 2023 Mar 8;45(3):2266-2283. doi: 10.3390/cimb45030146. Curr Issues Mol Biol. 2023. PMID: 36975516 Free PMC article. Review.
-
Biological Activity of Oleanolic Acid Derivatives HIMOXOL and Br-HIMOLID in Breast Cancer Cells Is Mediated by ER and EGFR.Int J Mol Sci. 2023 Mar 7;24(6):5099. doi: 10.3390/ijms24065099. Int J Mol Sci. 2023. PMID: 36982173 Free PMC article.
-
Principal Bioactive Properties of Oleanolic Acid, Its Derivatives, and Analogues.Molecules. 2024 Jul 12;29(14):3291. doi: 10.3390/molecules29143291. Molecules. 2024. PMID: 39064870 Free PMC article. Review.
References
-
- Miligy I.M., Toss M.S., Gorringe K.L., Lee A.H.S., Ellis I.O., Green A.R., Rakha E.A. The clinical and biological significance of HER2 overexpression in breast ductal carcinoma in situ: A large study from a single institution. Br. J. Cancer. 2019;120:1075–1082. doi: 10.1038/s41416-019-0436-3. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

