The role of SHP/REV-ERBα/CYP4A axis in the pathogenesis of alcohol-associated liver disease
- PMID: 34423788
- PMCID: PMC8410014
- DOI: 10.1172/jci.insight.140687
The role of SHP/REV-ERBα/CYP4A axis in the pathogenesis of alcohol-associated liver disease
Abstract
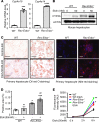
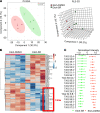
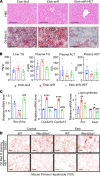
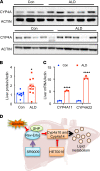
Alcohol-associated liver disease (ALD) represents a spectrum of histopathological changes, including alcoholic steatosis, steatohepatitis, and cirrhosis. One of the early responses to excessive alcohol consumption is lipid accumulation in the hepatocytes. Lipid ω-hydroxylation of medium- and long-chain fatty acid metabolized by the cytochrome P450 4A (CYP4A) family is an alternative pathway for fatty acid metabolism. The molecular mechanisms of CYP4A in ALD pathogenesis have not been elucidated. In this study, WT and Shp-/- mice were fed with a modified ethanol-binge, National Institute on Alcohol Abuse and Alcoholism model (10 days of ethanol feeding plus single binge). Liver tissues were collected every 6 hours for 24 hours and analyzed using RNA-Seq. The effects of REV-ERBα agonist (SR9009, 100 mg/kg/d) or CYP4A antagonist (HET0016, 5 mg/kg/d) in ethanol-fed mice were also evaluated. We found that hepatic Cyp4a10 and Cyp4a14 expression were significantly upregulated in WT mice, but not in Shp-/- mice, fed with ethanol. ChIP quantitative PCR and promoter assay revealed that REV-ERBα is the transcriptional repressor of Cyp4a10 and Cyp4a14. Rev-Erbα-/- hepatocytes had a marked induction of both Cyp4a genes and lipid accumulation. REV-ERBα agonist SR9009 or CYP4A antagonist HET0016 attenuated Cyp4a induction by ethanol and prevented alcohol-induced steatosis. Here, we have identified a role for the SHP/REV-ERBα/CYP4A axis in the pathogenesis of ALD. Our data also suggest REV-ERBα or CYP4A as the potential therapeutic targets for ALD.
Keywords: Fatty acid oxidation; Hepatology; Mouse models; Transcription.
Figures




Similar articles
-
REV-ERBα Activates C/EBP Homologous Protein to Control Small Heterodimer Partner-Mediated Oscillation of Alcoholic Fatty Liver.Am J Pathol. 2016 Nov;186(11):2909-2920. doi: 10.1016/j.ajpath.2016.07.014. Epub 2016 Sep 21. Am J Pathol. 2016. PMID: 27664470 Free PMC article.
-
Central role of PPARalpha-dependent hepatic lipid turnover in dietary steatohepatitis in mice.Hepatology. 2003 Jul;38(1):123-32. doi: 10.1053/jhep.2003.50307. Hepatology. 2003. PMID: 12829994
-
Pharmacological activation of REV-ERBα improves nonalcoholic steatohepatitis by regulating intestinal permeability.Metabolism. 2021 Jan;114:154409. doi: 10.1016/j.metabol.2020.154409. Epub 2020 Oct 21. Metabolism. 2021. PMID: 33096076
-
n-3 Polyunsaturated fatty acids for the management of alcoholic liver disease: A critical review.Crit Rev Food Sci Nutr. 2019;59(sup1):S116-S129. doi: 10.1080/10408398.2018.1544542. Epub 2018 Dec 22. Crit Rev Food Sci Nutr. 2019. PMID: 30580553 Review.
-
Dissecting the Rev-erbα Cistrome and the Mechanisms Controlling Circadian Transcription in Liver.Cold Spring Harb Symp Quant Biol. 2015;80:233-8. doi: 10.1101/sqb.2015.80.027508. Epub 2015 Sep 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26370410 Review.
Cited by
-
Single-Cell RNA Transcriptome Profiling of Liver Cells of Short-Term Alcoholic Liver Injury in Mice.Int J Mol Sci. 2023 Feb 22;24(5):4344. doi: 10.3390/ijms24054344. Int J Mol Sci. 2023. PMID: 36901774 Free PMC article.
-
Human umbilical cord mesenchymal stem cells attenuate diet-induced obesity and NASH-related fibrosis in mice.Heliyon. 2024 Feb 1;10(3):e25460. doi: 10.1016/j.heliyon.2024.e25460. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38356602 Free PMC article.
-
RNA-Seq Analysis of Protection against Chronic Alcohol Liver Injury by Rosa roxburghii Fruit Juice (Cili) in Mice.Nutrients. 2022 May 9;14(9):1974. doi: 10.3390/nu14091974. Nutrients. 2022. PMID: 35565941 Free PMC article.
-
Role of Circadian Transcription Factor Rev-Erb in Metabolism and Tissue Fibrosis.Int J Mol Sci. 2022 Oct 26;23(21):12954. doi: 10.3390/ijms232112954. Int J Mol Sci. 2022. PMID: 36361737 Free PMC article. Review.
-
Targeting NR1D1 in organ injury: challenges and prospects.Mil Med Res. 2023 Dec 11;10(1):62. doi: 10.1186/s40779-023-00495-3. Mil Med Res. 2023. PMID: 38072952 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

