Mass Spectrometry Imaging for Glycome in the Brain
- PMID: 34393728
- PMCID: PMC8358800
- DOI: 10.3389/fnana.2021.711955
Mass Spectrometry Imaging for Glycome in the Brain
Abstract
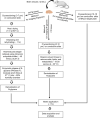
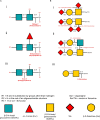
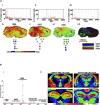
Glycans are diverse structured biomolecules that play crucial roles in various biological processes. Glycosylation, an enzymatic system through which various glycans are bound to proteins and lipids, is the most common and functionally crucial post-translational modification process. It is known to be associated with brain development, signal transduction, molecular trafficking, neurodegenerative disorders, psychopathologies, and brain cancers. Glycans in glycoproteins and glycolipids expressed in brain cells are involved in neuronal development, biological processes, and central nervous system maintenance. The composition and expression of glycans are known to change during those physiological processes. Therefore, imaging of glycans and the glycoconjugates in the brain regions has become a "hot" topic nowadays. Imaging techniques using lectins, antibodies, and chemical reporters are traditionally used for glycan detection. However, those techniques offer limited glycome detection. Mass spectrometry imaging (MSI) is an evolving field that combines mass spectrometry with histology allowing spatial and label-free visualization of molecules in the brain. In the last decades, several studies have employed MSI for glycome imaging in brain tissues. The current state of MSI uses on-tissue enzymatic digestion or chemical reaction to facilitate successful glycome imaging. Here, we reviewed the available literature that applied MSI techniques for glycome visualization and characterization in the brain. We also described the general methodologies for glycome MSI and discussed its potential use in the three-dimensional MSI in the brain.
Keywords: brain tissue; glycans; glycosylation; mass spectrometry imaging; three-dimensional MSI.
Copyright © 2021 Hasan, Mimi, Mamun, Islam, Waliullah, Nabi, Tamannaa, Kahyo and Setou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Understanding cellular glycan surfaces in the central nervous system.Biochem Soc Trans. 2019 Feb 28;47(1):89-100. doi: 10.1042/BST20180330. Epub 2018 Dec 17. Biochem Soc Trans. 2019. PMID: 30559272 Review.
-
Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues.Mol Imaging Biol. 2018 Dec;20(6):888-901. doi: 10.1007/s11307-018-1267-y. Mol Imaging Biol. 2018. PMID: 30167993 Free PMC article. Review.
-
A comprehensive glycome profiling of Huntington's disease transgenic mice.Biochim Biophys Acta. 2015 Sep;1850(9):1704-18. doi: 10.1016/j.bbagen.2015.04.006. Epub 2015 Apr 20. Biochim Biophys Acta. 2015. PMID: 25907331
-
N-Glycan matrix-assisted laser desorption/ionization mass spectrometry imaging protocol for formalin-fixed paraffin-embedded tissues.Rapid Commun Mass Spectrom. 2017 May 30;31(10):825-841. doi: 10.1002/rcm.7845. Rapid Commun Mass Spectrom. 2017. PMID: 28271569
-
Altered N-linked glycosylation in endometrial cancer.Anal Bioanal Chem. 2021 Apr;413(10):2721-2733. doi: 10.1007/s00216-020-03039-z. Epub 2020 Nov 21. Anal Bioanal Chem. 2021. PMID: 33222001
Cited by
-
Theranostic Applications of Glycosaminoglycans in Metastatic Renal Cell Carcinoma.Cancers (Basel). 2022 Dec 30;15(1):266. doi: 10.3390/cancers15010266. Cancers (Basel). 2022. PMID: 36612261 Free PMC article. Review.
-
An N-glycome tissue atlas of 15 human normal and cancer tissue types determined by MALDI-imaging mass spectrometry.Sci Rep. 2024 Jan 4;14(1):489. doi: 10.1038/s41598-023-50957-w. Sci Rep. 2024. PMID: 38177192 Free PMC article.
-
Changes in tissue protein N-glycosylation and associated molecular signature occur in the human Parkinsonian brain in a region-specific manner.PNAS Nexus. 2023 Dec 25;3(1):pgad439. doi: 10.1093/pnasnexus/pgad439. eCollection 2024 Jan. PNAS Nexus. 2023. PMID: 38178977 Free PMC article.
-
N-glycans show distinct spatial distribution in mouse brain.Glycobiology. 2023 Dec 25;33(11):935-942. doi: 10.1093/glycob/cwad077. Glycobiology. 2023. PMID: 37792804 Free PMC article.
-
Novel mimetic tissue standards for precise quantitative mass spectrometry imaging of drug and neurotransmitter concentrations in rat brain tissues.Anal Bioanal Chem. 2024 Nov;416(26):5579-5593. doi: 10.1007/s00216-024-05477-5. Epub 2024 Aug 10. Anal Bioanal Chem. 2024. PMID: 39126505 Free PMC article.
References
-
- Brandenburg K., Holst O. (2015). Glycolipids: Distribution and Biological Function. Chichester, UK: John Wiley & Sons, Ltd, 1–10. 10.1002/9780470015902.a0001427.pub3 - DOI
Publication types
LinkOut - more resources
Full Text Sources

