The association of epigenetic clocks in brain tissue with brain pathologies and common aging phenotypes
- PMID: 34153464
- PMCID: PMC8373772
- DOI: 10.1016/j.nbd.2021.105428
The association of epigenetic clocks in brain tissue with brain pathologies and common aging phenotypes
Abstract
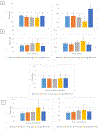
Epigenetic clocks are calculated by combining DNA methylation states across select CpG sites to estimate biologic age, and have been noted as the most successful markers of biologic aging to date. Yet, limited research has considered epigenetic clocks calculated in brain tissue. We used DNA methylation states in dorsolateral prefrontal cortex specimens from 721 older participants of the Religious Orders Study and Rush Memory and Aging Project, to calculate DNA methylation age using four established epigenetic clocks: Hannum, Horvath, PhenoAge, GrimAge, and a new Cortical clock. The four established clocks were trained in blood samples (Hannum, PhenoAge, GrimAge) or using 51 human tissue and cell types (Horvath); the recent Cortical clock is the first trained in postmortem cortical tissue. Participants were recruited beginning in 1994 (Religious Orders Study) and 1997 (Memory and Aging Project), and followed annually with questionnaires and clinical evaluations; brain specimens were obtained for 80-90% of participants. Mean age at death was 88.0 (SD 6.7) years. We used linear regression, logistic regression, and linear mixed models, to examine relations of epigenetic clock ages to neuropathologic and clinical aging phenotypes, controlling for chronologic age, sex, education, and depressive symptomatology. Hannum, Horvath, PhenoAge and Cortical clock ages were related to pathologic diagnosis of Alzheimer's disease (AD), as well as to Aβ load (a hallmark pathology of Alzheimer's disease). However, associations were substantially stronger for the Cortical than other clocks; for example, each standard deviation (SD) increase in Hannum, Horvath, and PhenoAge clock age was related to approximately 30% greater likelihood of pathologic AD (all p < 0.05), while each SD increase in Cortical age was related to 90% greater likelihood of pathologic AD (odds ratio = 1.91, 95% confidence interval 1.38, 2.62). Moreover, Cortical age was significantly related to other AD pathology (eg, mean tau tangle density, p = 0.003), and to odds of neocortical Lewy body pathology (for each SD increase in Cortical age, odds ratio = 2.00, 95% confidence 1.27, 3.17), although no clocks were related to cerebrovascular neuropathology. Cortical age was the only epigenetic clock significantly associated with the clinical phenotypes examined, from dementia to cognitive decline (5 specific cognitive systems, and a global cognitive measure averaging 17 tasks) to Parkinsonian signs. Overall, our findings provide evidence of the critical necessity for bespoke clocks of brain aging for advancing research to understand, and eventually prevent, neurodegenerative diseases of aging.
Keywords: Aging; Dementia; Epidemiology; Epigenetics; Neuropathology.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Competing Interests:
The authors have no competing interests to declare.
Figures

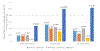
Similar articles
-
Characteristics of Epigenetic Clocks Across Blood and Brain Tissue in Older Women and Men.Front Neurosci. 2021 Jan 7;14:555307. doi: 10.3389/fnins.2020.555307. eCollection 2020. Front Neurosci. 2021. PMID: 33488342 Free PMC article.
-
The influences of DNA methylation and epigenetic clocks, on metabolic disease, in middle-aged Koreans.Clin Epigenetics. 2020 Oct 15;12(1):148. doi: 10.1186/s13148-020-00936-z. Clin Epigenetics. 2020. PMID: 33059731 Free PMC article.
-
Association of Neighborhood Deprivation With Epigenetic Aging Using 4 Clock Metrics.JAMA Netw Open. 2020 Nov 2;3(11):e2024329. doi: 10.1001/jamanetworkopen.2020.24329. JAMA Netw Open. 2020. PMID: 33146735 Free PMC article.
-
A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration.Ageing Res Rev. 2021 Aug;69:101348. doi: 10.1016/j.arr.2021.101348. Epub 2021 Apr 28. Ageing Res Rev. 2021. PMID: 33930583 Review.
-
Epigenetic Clocks: In Aging-Related and Complex Diseases.Cytogenet Genome Res. 2023;163(5-6):247-256. doi: 10.1159/000534561. Epub 2023 Oct 28. Cytogenet Genome Res. 2023. PMID: 37899027 Review.
Cited by
-
Author Response: Estrogen Receptor Genes, Cognitive Decline, and Alzheimer Disease.Neurology. 2023 Oct 3;101(14):634. doi: 10.1212/WNL.0000000000207842. Neurology. 2023. PMID: 37783504 Free PMC article. No abstract available.
-
Evaluating genomic signatures of aging in brain tissue as it relates to Alzheimer's disease.Sci Rep. 2023 Sep 7;13(1):14747. doi: 10.1038/s41598-023-41400-1. Sci Rep. 2023. PMID: 37679407 Free PMC article.
-
Entorhinal cortex epigenome-wide association study highlights four novel loci showing differential methylation in Alzheimer's disease.Alzheimers Res Ther. 2023 May 6;15(1):92. doi: 10.1186/s13195-023-01232-7. Alzheimers Res Ther. 2023. PMID: 37149695 Free PMC article.
-
Histopathologic brain age estimation via multiple instance learning.Acta Neuropathol. 2023 Dec;146(6):785-802. doi: 10.1007/s00401-023-02636-3. Epub 2023 Oct 10. Acta Neuropathol. 2023. PMID: 37815677 Free PMC article.
-
Epigenetic modifications of DNA and RNA in Alzheimer's disease.Front Mol Neurosci. 2024 Apr 25;17:1398026. doi: 10.3389/fnmol.2024.1398026. eCollection 2024. Front Mol Neurosci. 2024. PMID: 38726308 Free PMC article. Review.
References
-
- Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, Whitsel EA, Assimes TL, Ferrucci L, Horvath S. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019January21;11(2):303–327. doi: 10.18632/aging.101684. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

