Unlike LGR4, LGR5 potentiates Wnt-β-catenin signaling without sequestering E3 ligases
- PMID: 33262293
- PMCID: PMC7905944
- DOI: 10.1126/scisignal.aaz4051
Unlike LGR4, LGR5 potentiates Wnt-β-catenin signaling without sequestering E3 ligases
Abstract
LGR4 and LGR5 encode two homologous receptors with critical, yet distinct, roles in organ development and adult stem cell survival. Both receptors are coexpressed in intestinal crypt stem cells, bind to R-spondins (RSPOs) with high affinity, and potentiate Wnt-β-catenin signaling, presumably by the same mechanism: forming RSPO-bridged complexes with the E3 ligases RNF43 and ZNRF3 to inhibit ubiquitylation of Wnt receptors. However, direct evidence for RSPO-bound, full-length LGR5 interacting with these E3 ligases in whole cells has not been reported, and only LGR4 is essential for the self-renewal of intestinal stem cells. Here, we examined the mechanisms of action of LGR4 and LGR5 in parallel using coimmunoprecipitation, proximity ligation, competition binding, and time-resolved FRET assays in whole cells. Full-length LGR4 formed a tight complex with ZNRF3 and RNF43 even without RSPO, whereas LGR5 did not interact with either E3 ligase with or without RSPO. Domain-swapping experiments with LGR4 and LGR5 revealed that the seven-transmembrane domain of LGR4 conferred interaction with the E3 ligases. Native LGR4 and LGR5 existed as dimers on the cell surface, and LGR5 interacted with both FZD and LRP6 of the Wnt signalosome to enhance LRP6 phosphorylation and potentiate Wnt-β-catenin signaling. These findings provide a molecular basis for the weaker activity of LGR5 in the potentiation of Wnt signaling that may underlie the distinct roles of LGR4 and LGR5 in organ development, as well as the self-renewal and fitness of adult stem cells.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


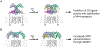
Similar articles
-
LGR4 and LGR5 form distinct homodimers that only LGR4 complexes with RNF43/ZNRF3 to provide high affinity binding of R-spondin ligands.Sci Rep. 2023 Jul 4;13(1):10796. doi: 10.1038/s41598-023-37856-w. Sci Rep. 2023. PMID: 37402772 Free PMC article.
-
RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling.Proc Natl Acad Sci U S A. 2014 Apr 1;111(13):E1221-9. doi: 10.1073/pnas.1323106111. Epub 2014 Mar 17. Proc Natl Acad Sci U S A. 2014. PMID: 24639526 Free PMC article.
-
Differential activities and mechanisms of the four R-spondins in potentiating Wnt/β-catenin signaling.J Biol Chem. 2018 Jun 22;293(25):9759-9769. doi: 10.1074/jbc.RA118.002743. Epub 2018 May 11. J Biol Chem. 2018. PMID: 29752411 Free PMC article.
-
The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength.Genes Dev. 2014 Feb 15;28(4):305-16. doi: 10.1101/gad.235473.113. Genes Dev. 2014. PMID: 24532711 Free PMC article. Review.
-
The RSPO-LGR4/5-ZNRF3/RNF43 module in liver homeostasis, regeneration, and disease.Hepatology. 2022 Sep;76(3):888-899. doi: 10.1002/hep.32328. Epub 2022 Feb 20. Hepatology. 2022. PMID: 35006616 Review.
Cited by
-
Single-cell transcriptomics shows dose-dependent disruption of hepatic zonation by TCDD in mice.Toxicol Sci. 2023 Jan 31;191(1):135-148. doi: 10.1093/toxsci/kfac109. Toxicol Sci. 2023. PMID: 36222588 Free PMC article.
-
LGR5 as a Therapeutic Target of Antibody-Functionalized Biomimetic Magnetoliposomes for Colon Cancer Therapy.Int J Nanomedicine. 2024 Feb 23;19:1843-1865. doi: 10.2147/IJN.S440881. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38414530 Free PMC article.
-
Drug Conjugates of Antagonistic R-Spondin 4 Mutant for Simultaneous Targeting of Leucine-Rich Repeat-Containing G Protein-Coupled Receptors 4/5/6 for Cancer Treatment.J Med Chem. 2021 Sep 9;64(17):12572-12581. doi: 10.1021/acs.jmedchem.1c00395. Epub 2021 Aug 18. J Med Chem. 2021. PMID: 34406767 Free PMC article.
-
LGR4, a G Protein-Coupled Receptor With a Systemic Role: From Development to Metabolic Regulation.Front Endocrinol (Lausanne). 2022 May 30;13:867001. doi: 10.3389/fendo.2022.867001. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35707461 Free PMC article. Review.
-
G protein-coupled receptor-targeting antibody-drug conjugates: Current status and future directions.Cancer Lett. 2023 Jun 28;564:216191. doi: 10.1016/j.canlet.2023.216191. Epub 2023 Apr 25. Cancer Lett. 2023. PMID: 37100113 Free PMC article. Review.
References
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H, Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007); published online EpubOct 25 (10.1038/nature06196). - DOI - PubMed
-
- McDonald T, Wang R, Bailey W, Xie G, Chen F, Caskey CT, Liu Q, Identification and cloning of an orphan G protein-coupled receptor of the glycoprotein hormone receptor subfamily. Biochem Biophys Res Commun 247, 266–270 (1998); published online EpubJun 18 (S0006–291X(98)98774–5 [pii]10.1006/bbrc.1998.8774). - DOI - PubMed
-
- Hsu SY, Liang SG, Hsueh AJ, Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol Endocrinol 12, 1830–1845 (1998); published online EpubDec (10.1210/mend.12.12.0211) - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

