De Novo Damaging Variants, Clinical Phenotypes, and Post-Operative Outcomes in Congenital Heart Disease
- PMID: 32812804
- PMCID: PMC7439931
- DOI: 10.1161/CIRCGEN.119.002836
De Novo Damaging Variants, Clinical Phenotypes, and Post-Operative Outcomes in Congenital Heart Disease
Abstract
Background: De novo genic and copy number variants are enriched in patients with congenital heart disease, particularly those with extra-cardiac anomalies. The impact of de novo damaging variants on outcomes following cardiac repair is unknown.
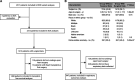
Methods: We studied 2517 patients with congenital heart disease who had undergone whole-exome sequencing as part of the CHD GENES study (Congenital Heart Disease Genetic Network).
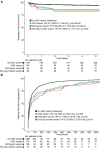
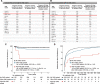
Results: Two hundred ninety-four patients (11.7%) had clinically significant de novo variants. Patients with de novo damaging variants were 2.4 times more likely to have extra-cardiac anomalies (P=5.63×10-12). In 1268 patients (50.4%) who had surgical data available and underwent open-heart surgery exclusive of heart transplantation as their first operation, we analyzed transplant-free survival following the first operation. Median follow-up was 2.65 years. De novo variants were associated with worse transplant-free survival (hazard ratio, 3.51; P=5.33×10-04) and longer times to final extubation (hazard ratio, 0.74; P=0.005). As de novo variants had a significant interaction with extra-cardiac anomalies for transplant-free survival (P=0.003), de novo variants conveyed no additional risk for transplant-free survival for patients with these anomalies (adjusted hazard ratio, 1.96; P=0.06). By contrast, de novo variants in patients without extra-cardiac anomalies were associated with worse transplant-free survival during follow-up (hazard ratio, 11.21; P=1.61×10-05) than that of patients with no de novo variants. Using agnostic machine-learning algorithms, we identified de novo copy number variants at 15q25.2 and 15q11.2 as being associated with worse transplant-free survival and 15q25.2, 22q11.21, and 3p25.2 as being associated with prolonged time to final extubation.
Conclusions: In patients with congenital heart disease undergoing open-heart surgery, de novo variants were associated with worse transplant-free survival and longer times on the ventilator. De novo variants were most strongly associated with adverse outcomes among patients without extra-cardiac anomalies, suggesting a benefit for preoperative genetic testing even when genetic abnormalities are not suspected during routine clinical practice. Registration: URL: https://www.clinicaltrials.gov. Unique identifier: NCT01196182.
Keywords: congenital heart disease; genomics; heart transplantation; mortality; survival.
Conflict of interest statement
None.
Figures

Similar articles
-
In-Depth Genomic Analysis: The New Challenge in Congenital Heart Disease.Int J Mol Sci. 2024 Feb 1;25(3):1734. doi: 10.3390/ijms25031734. Int J Mol Sci. 2024. PMID: 38339013 Free PMC article. Review.
-
Burden of potentially pathologic copy number variants is higher in children with isolated congenital heart disease and significantly impairs covariate-adjusted transplant-free survival.J Thorac Cardiovasc Surg. 2016 Apr;151(4):1147-51.e4. doi: 10.1016/j.jtcvs.2015.09.136. Epub 2015 Nov 10. J Thorac Cardiovasc Surg. 2016. PMID: 26704054 Free PMC article.
-
De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders.PLoS Genet. 2018 Dec 10;14(12):e1007822. doi: 10.1371/journal.pgen.1007822. eCollection 2018 Dec. PLoS Genet. 2018. PMID: 30532227 Free PMC article.
-
The contribution of de novo and rare inherited copy number changes to congenital heart disease in an unselected sample of children with conotruncal defects or hypoplastic left heart disease.Hum Genet. 2014 Jan;133(1):11-27. doi: 10.1007/s00439-013-1353-9. Epub 2013 Aug 25. Hum Genet. 2014. PMID: 23979609 Free PMC article.
-
Trends and outcomes in transplantation for complex congenital heart disease: 1984 to 2004.Ann Thorac Surg. 2004 Oct;78(4):1352-61; discussion 1352-61. doi: 10.1016/j.athoracsur.2004.04.012. Ann Thorac Surg. 2004. PMID: 15464499 Review.
Cited by
-
Early ascertainment of genetic diagnoses clarifies impact on medium-term survival following neonatal congenital heart surgery.J Clin Invest. 2024 Jul 30;134(18):e180098. doi: 10.1172/JCI180098. J Clin Invest. 2024. PMID: 39078715 Free PMC article. No abstract available.
-
How Will Artificial Intelligence Shape the Future of Decision-Making in Congenital Heart Disease?J Clin Med. 2024 May 20;13(10):2996. doi: 10.3390/jcm13102996. J Clin Med. 2024. PMID: 38792537 Free PMC article. Review.
-
Using novel data linkage of congenital heart disease biobank data with administrative health data to identify cardiovascular outcomes to inform genomic analysis.Int J Popul Data Sci. 2023 Nov 14;8(1):2150. doi: 10.23889/ijpds.v8i1.2150. eCollection 2023. Int J Popul Data Sci. 2023. PMID: 38414539 Free PMC article.
-
In-Depth Genomic Analysis: The New Challenge in Congenital Heart Disease.Int J Mol Sci. 2024 Feb 1;25(3):1734. doi: 10.3390/ijms25031734. Int J Mol Sci. 2024. PMID: 38339013 Free PMC article. Review.
-
Clinically Relevant Genetic Considerations for Patients With Tetralogy of Fallot.CJC Pediatr Congenit Heart Dis. 2023 Oct 10;2(6Part A):426-439. doi: 10.1016/j.cjcpc.2023.10.002. eCollection 2023 Dec. CJC Pediatr Congenit Heart Dis. 2023. PMID: 38161665 Free PMC article. Review.
References
-
- van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. - PubMed
-
- Matthews TJ, MacDorman MF. Infant mortality statistics from the 2010 period linked birth/infant death data set. Natl Vital Stat Rep. 2013;62:1–26. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
- T32 HL007572/HL/NHLBI NIH HHS/United States
- U01 HL098162/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01 HL098153/HL/NHLBI NIH HHS/United States
- U01 HL153009/HL/NHLBI NIH HHS/United States
- U54 HG006504/HG/NHGRI NIH HHS/United States
- UL1 TR000003/TR/NCATS NIH HHS/United States
- UM1 HL128761/HL/NHLBI NIH HHS/United States
- U01 HL098123/HL/NHLBI NIH HHS/United States
- UM1 HL128711/HL/NHLBI NIH HHS/United States
- UM1 HL098147/HL/NHLBI NIH HHS/United States
- U01 HL098147/HL/NHLBI NIH HHS/United States
- U01 HL098163/HL/NHLBI NIH HHS/United States
- UM1 HL098162/HL/NHLBI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- U01 HL098188/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

