AAV-Genome Population Sequencing of Vectors Packaging CRISPR Components Reveals Design-Influenced Heterogeneity
- PMID: 32775498
- PMCID: PMC7397707
- DOI: 10.1016/j.omtm.2020.07.007
AAV-Genome Population Sequencing of Vectors Packaging CRISPR Components Reveals Design-Influenced Heterogeneity
Abstract
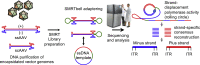
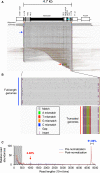
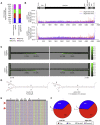
The gene therapy field has been galvanized by two technologies that have revolutionized treating genetic diseases: vectors based on adeno-associated viruses (AAVs), and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas gene-editing tools. When combined into one platform, these safe and broadly tropic biotherapies can be engineered to target any region in the human genome to correct genetic flaws. Unfortunately, few investigations into the design compatibility of CRISPR components in AAV vectors exist. Using AAV-genome population sequencing (AAV-GPseq), we previously found that self-complementary AAV vector designs with strong DNA secondary structures can cause a high degree of truncation events, impacting production and vector efficacy. We hypothesized that the single-guide RNA (sgRNA) scaffold, which contains several loop regions, may also compromise vector integrity. We have therefore advanced the AAV-GPseq method to also interrogate single-strand AAV vectors to investigate whether vector genomes carrying Cas9-sgRNA cassettes can cause truncation events. We found that on their own, sgRNA sequences do not produce a high degree of truncation events. However, we demonstrate that vector genome designs that carry dual sgRNA expression cassettes in tail-to-tail configurations lead to truncations. In addition, we revealed that heterogeneity in inverted terminal repeat sequences in the form of regional deletions inherent to certain AAV vector plasmids can be interrogated.
Keywords: AAV-genome population sequencing; CRISPR-Cas; ITR; adeno-associated virus; gene therapy vectors; real-time sequencing; scAAV; sgRNA; single molecule; ssAAV.
Figures



Similar articles
-
Adeno-associated Virus Genome Population Sequencing Achieves Full Vector Genome Resolution and Reveals Human-Vector Chimeras.Mol Ther Methods Clin Dev. 2018 Feb 13;9:130-141. doi: 10.1016/j.omtm.2018.02.002. eCollection 2018 Jun 15. Mol Ther Methods Clin Dev. 2018. PMID: 29766023 Free PMC article.
-
A Comprehensive Study of the Effects by Sequence Truncation within Inverted Terminal Repeats (ITRs) on the Productivity, Genome Packaging, and Potency of AAV Vectors.Microorganisms. 2024 Feb 1;12(2):310. doi: 10.3390/microorganisms12020310. Microorganisms. 2024. PMID: 38399714 Free PMC article.
-
Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes.J Virol. 2005 Jan;79(1):364-79. doi: 10.1128/JVI.79.1.364-379.2005. J Virol. 2005. PMID: 15596830 Free PMC article.
-
CRISPR Systems Suitable for Single AAV Vector Delivery.Curr Gene Ther. 2022;22(1):1-14. doi: 10.2174/1566523221666211006120355. Curr Gene Ther. 2022. PMID: 34620062 Review.
-
In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease.F1000Res. 2017 Dec 20;6:2153. doi: 10.12688/f1000research.11243.1. eCollection 2017. F1000Res. 2017. PMID: 29333255 Free PMC article. Review.
Cited by
-
Preventing packaging of translatable P5-associated DNA contaminants in recombinant AAV vector preps.Mol Ther Methods Clin Dev. 2022 Jan 19;24:280-291. doi: 10.1016/j.omtm.2022.01.008. eCollection 2022 Mar 10. Mol Ther Methods Clin Dev. 2022. PMID: 35211640 Free PMC article.
-
Strategies to improve safety profile of AAV vectors.Front Mol Med. 2022 Nov 1;2:1054069. doi: 10.3389/fmmed.2022.1054069. eCollection 2022. Front Mol Med. 2022. PMID: 39086961 Free PMC article. Review.
-
Stowaways in the cargo: Contaminating nucleic acids in rAAV preparations for gene therapy.Mol Ther. 2023 Oct 4;31(10):2826-2838. doi: 10.1016/j.ymthe.2023.07.025. Epub 2023 Aug 2. Mol Ther. 2023. PMID: 37533254 Free PMC article. Review.
-
AAVolve: Concatenated long-read deep sequencing enables whole capsid tracking during shuffled AAV library selection.Mol Ther Methods Clin Dev. 2024 Oct 4;32(4):101351. doi: 10.1016/j.omtm.2024.101351. eCollection 2024 Dec 12. Mol Ther Methods Clin Dev. 2024. PMID: 39498467 Free PMC article.
-
DNA read count calibration for single-molecule, long-read sequencing.Sci Rep. 2022 Nov 1;12(1):17257. doi: 10.1038/s41598-022-21606-5. Sci Rep. 2022. PMID: 36319642 Free PMC article.
References
-
- Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

