Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis
- PMID: 32661339
- PMCID: PMC7461628
- DOI: 10.1038/s41556-020-0542-8
Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis
Abstract
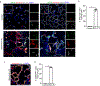
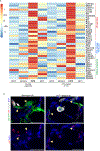
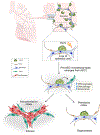
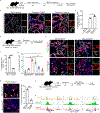
Stem cells undergo dynamic changes in response to injury to regenerate lost cells. However, the identity of transitional states and the mechanisms that drive their trajectories remain understudied. Using lung organoids, multiple in vivo repair models, single-cell transcriptomics and lineage tracing, we find that alveolar type-2 epithelial cells undergoing differentiation into type-1 cells acquire pre-alveolar type-1 transitional cell state (PATS) en route to terminal maturation. Transitional cells undergo extensive stretching during differentiation, making them vulnerable to DNA damage. Cells in the PATS show an enrichment of TP53, TGFβ, DNA-damage-response signalling and cellular senescence. Gain and loss of function as well as genomic binding assays revealed a direct transcriptional control of PATS by TP53 signalling. Notably, accumulation of PATS-like cells in human fibrotic lungs was observed, suggesting persistence of the transitional state in fibrosis. Our study thus implicates a transient state associated with senescence in normal epithelial tissue repair and its abnormal persistence in disease conditions.
Conflict of interest statement
Competing interests
The authors declare the following competing interests: A provisional patent application related to this work has been filed. Y.K., A.T., A.K., and P.R.T. are listed as co-inventors on this application; P.R.T. serves as a consultant for Cellarity Inc. not related to this work.
Figures













Comment in
-
A transitional stem cell state in the lung.Nat Cell Biol. 2020 Sep;22(9):1025-1026. doi: 10.1038/s41556-020-0561-5. Nat Cell Biol. 2020. PMID: 32778743 No abstract available.
Similar articles
-
Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis.Nat Commun. 2020 Jul 16;11(1):3559. doi: 10.1038/s41467-020-17358-3. Nat Commun. 2020. PMID: 32678092 Free PMC article.
-
p53 governs an AT1 differentiation programme in lung cancer suppression.Nature. 2023 Jul;619(7971):851-859. doi: 10.1038/s41586-023-06253-8. Epub 2023 Jul 19. Nature. 2023. PMID: 37468633 Free PMC article.
-
Disease modeling of pulmonary fibrosis using human pluripotent stem cell-derived alveolar organoids.Stem Cell Reports. 2021 Dec 14;16(12):2973-2987. doi: 10.1016/j.stemcr.2021.10.015. Epub 2021 Nov 18. Stem Cell Reports. 2021. PMID: 34798066 Free PMC article.
-
Regeneration-Associated Transitional State Cells in Pulmonary Fibrosis.Int J Mol Sci. 2022 Jun 17;23(12):6757. doi: 10.3390/ijms23126757. Int J Mol Sci. 2022. PMID: 35743199 Free PMC article. Review.
-
Alveolar wars: The rise of in vitro models to understand human lung alveolar maintenance, regeneration, and disease.Stem Cells Transl Med. 2020 Aug;9(8):867-881. doi: 10.1002/sctm.19-0433. Epub 2020 Apr 9. Stem Cells Transl Med. 2020. PMID: 32272001 Free PMC article. Review.
Cited by
-
PCLAF-DREAM drives alveolar cell plasticity for lung regeneration.Nat Commun. 2024 Oct 24;15(1):9169. doi: 10.1038/s41467-024-53330-1. Nat Commun. 2024. PMID: 39448571 Free PMC article.
-
Heterogeneous groups of alveolar type II cells in lung homeostasis and repair.Am J Physiol Cell Physiol. 2020 Dec 1;319(6):C991-C996. doi: 10.1152/ajpcell.00341.2020. Epub 2020 Sep 9. Am J Physiol Cell Physiol. 2020. PMID: 32903031 Free PMC article. Review.
-
Contributions of alveolar epithelial cell quality control to pulmonary fibrosis.J Clin Invest. 2020 Oct 1;130(10):5088-5099. doi: 10.1172/JCI139519. J Clin Invest. 2020. PMID: 32870817 Free PMC article. Review.
-
The potential for OGG1 inhibition to be a therapeutic strategy for pulmonary diseases.Expert Opin Ther Targets. 2024 Mar;28(3):117-130. doi: 10.1080/14728222.2024.2317900. Epub 2024 Feb 14. Expert Opin Ther Targets. 2024. PMID: 38344773 Review.
-
Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration.Cell Stem Cell. 2020 Sep 3;27(3):366-382.e7. doi: 10.1016/j.stem.2020.06.020. Epub 2020 Aug 3. Cell Stem Cell. 2020. PMID: 32750316 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

