Regulating tumor suppressor genes: post-translational modifications
- PMID: 32532965
- PMCID: PMC7293209
- DOI: 10.1038/s41392-020-0196-9
Regulating tumor suppressor genes: post-translational modifications
Abstract
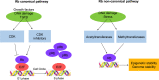
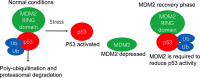
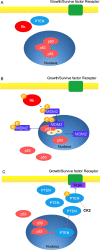
Tumor suppressor genes cooperate with each other in tumors. Three important tumor suppressor proteins, retinoblastoma (Rb), p53, phosphatase, and tensin homolog deleted on chromosome ten (PTEN) are functionally associated and they regulated by post-translational modification (PTMs) as well. PTMs include phosphorylation, SUMOylation, acetylation, and other novel modifications becoming growing appreciated. Because most of PTMs are reversible, normal cells use them as a switch to control the state of cells being the resting or proliferating, and PTMs also involve in cell survival and cell cycle, which may lead to abnormal proliferation and tumorigenesis. Although a lot of studies focus on the importance of each kind of PTM, further discoveries shows that tumor suppressor genes (TSGs) form a complex "network" by the interaction of modification. Recently, there are several promising strategies for TSGs for they change more frequently than carcinogenic genes in cancers. We here review the necessity, characteristics, and mechanisms of each kind of post-translational modification on Rb, p53, PTEN, and its influence on the precise and selective function. We also discuss the current antitumoral therapies of Rb, p53 and PTEN as predictive, prognostic, and therapeutic target in cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Evasion of anti-growth signaling: A key step in tumorigenesis and potential target for treatment and prophylaxis by natural compounds.Semin Cancer Biol. 2015 Dec;35 Suppl:S55-S77. doi: 10.1016/j.semcancer.2015.02.005. Epub 2015 Mar 6. Semin Cancer Biol. 2015. PMID: 25749195 Free PMC article. Review.
-
An in vitro system to characterize prostate cancer progression identified signaling required for self-renewal.Mol Carcinog. 2016 Dec;55(12):1974-1989. doi: 10.1002/mc.22444. Epub 2015 Dec 1. Mol Carcinog. 2016. PMID: 26621780
-
Decoding PTEN: from biological functions to signaling pathways in tumors.Mol Biol Rep. 2024 Oct 24;51(1):1089. doi: 10.1007/s11033-024-10049-y. Mol Biol Rep. 2024. PMID: 39446204 Review.
-
Immunohistochemical and molecular analysis of p53, RB, and PTEN in malignant peripheral nerve sheath tumors.Virchows Arch. 2002 Jun;440(6):610-5. doi: 10.1007/s00428-001-0550-4. Epub 2001 Nov 16. Virchows Arch. 2002. PMID: 12070601
-
p53 modifications: exquisite decorations of the powerful guardian.J Mol Cell Biol. 2019 Jul 19;11(7):564-577. doi: 10.1093/jmcb/mjz060. J Mol Cell Biol. 2019. PMID: 31282934 Free PMC article. Review.
Cited by
-
Krüppel-like factor 4 promotes survival and expansion in acute myeloid leukemia cells.Oncotarget. 2021 Feb 16;12(4):255-267. doi: 10.18632/oncotarget.27878. eCollection 2021 Feb 16. Oncotarget. 2021. PMID: 33659038 Free PMC article.
-
Comprehensive characterization of ubiquitinome of human colorectal cancer and identification of potential survival-related ubiquitination.J Transl Med. 2022 Oct 2;20(1):445. doi: 10.1186/s12967-022-03645-8. J Transl Med. 2022. PMID: 36184622 Free PMC article.
-
Circular RNA Controls Tumor Occurrence and Development via Cell Cycle Regulation.Onco Targets Ther. 2022 Sep 15;15:993-1009. doi: 10.2147/OTT.S371629. eCollection 2022. Onco Targets Ther. 2022. PMID: 36134387 Free PMC article. Review.
-
Pathological implication of protein post-translational modifications in cancer.Mol Aspects Med. 2022 Aug;86:101097. doi: 10.1016/j.mam.2022.101097. Epub 2022 Apr 7. Mol Aspects Med. 2022. PMID: 35400524 Free PMC article. Review.
-
A Bibliometric Analysis Based on Web of Science: Current Perspectives and Potential Trends of SMAD7 in Oncology.Front Cell Dev Biol. 2022 Feb 18;9:712732. doi: 10.3389/fcell.2021.712732. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35252215 Free PMC article.
References
-
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. - PubMed
-
- Bishop JM, et al. Origin and function of avian retrovirus transforming genes. Cold Spring Harb. Symp. Quant. Biol. 1980;44:919–930. - PubMed
-
- Parris GE. The cell clone ecology hypothesis and the cell fusion model of cancer progression and metastasis: history and experimental support. Med. Hypotheses. 2006;66:76–83. - PubMed
-
- Bashyam MD, Animireddy S, Bala P, Naz A, George SA. The Yin and Yang of cancer genes. Gene. 2019;704:121–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

