PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function
- PMID: 32451509
- PMCID: PMC7405943
- DOI: 10.1038/s41589-020-0553-6
PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function
Abstract
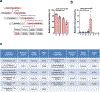
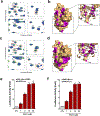
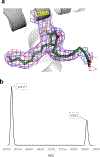
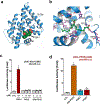
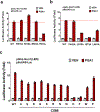
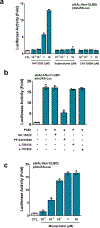
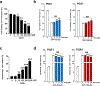
The orphan nuclear receptor Nurr1 is critical for the development, maintenance and protection of midbrain dopaminergic (mDA) neurons. Here we show that prostaglandin E1 (PGE1) and its dehydrated metabolite, PGA1, directly interact with the ligand-binding domain (LBD) of Nurr1 and stimulate its transcriptional function. We also report the crystallographic structure of Nurr1-LBD bound to PGA1 at 2.05 Å resolution. PGA1 couples covalently to Nurr1-LBD by forming a Michael adduct with Cys566, and induces notable conformational changes, including a 21° shift of the activation function-2 helix (H12) away from the protein core. Furthermore, PGE1/PGA1 exhibit neuroprotective effects in a Nurr1-dependent manner, prominently enhance expression of Nurr1 target genes in mDA neurons and improve motor deficits in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-lesioned mouse models of Parkinson's disease. Based on these results, we propose that PGE1/PGA1 represent native ligands of Nurr1 and can exert neuroprotective effects on mDA neurons, via activation of Nurr1's transcriptional function.
Figures

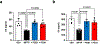



Similar articles
-
Prostaglandin A2 Interacts with Nurr1 and Ameliorates Behavioral Deficits in Parkinson's Disease Fly Model.Neuromolecular Med. 2022 Dec;24(4):469-478. doi: 10.1007/s12017-022-08712-3. Epub 2022 Apr 28. Neuromolecular Med. 2022. PMID: 35482177
-
Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.Cell Chem Biol. 2019 May 16;26(5):674-685.e6. doi: 10.1016/j.chembiol.2019.02.002. Epub 2019 Mar 7. Cell Chem Biol. 2019. PMID: 30853418 Free PMC article.
-
Nurr1 and Retinoid X Receptor Ligands Stimulate Ret Signaling in Dopamine Neurons and Can Alleviate α-Synuclein Disrupted Gene Expression.J Neurosci. 2015 Oct 21;35(42):14370-85. doi: 10.1523/JNEUROSCI.1155-15.2015. J Neurosci. 2015. PMID: 26490873 Free PMC article.
-
Potent synthetic and endogenous ligands for the adopted orphan nuclear receptor Nurr1.Exp Mol Med. 2021 Jan;53(1):19-29. doi: 10.1038/s12276-021-00555-5. Epub 2021 Jan 21. Exp Mol Med. 2021. PMID: 33479411 Free PMC article. Review.
-
Medicinal Chemistry and Chemical Biology of Nurr1 Modulators: An Emerging Strategy in Neurodegeneration.J Med Chem. 2022 Jul 28;65(14):9548-9563. doi: 10.1021/acs.jmedchem.2c00585. Epub 2022 Jul 7. J Med Chem. 2022. PMID: 35797147 Review.
Cited by
-
Developmental pathways linked to the vulnerability of adult midbrain dopaminergic neurons to neurodegeneration.Front Mol Neurosci. 2022 Dec 22;15:1071731. doi: 10.3389/fnmol.2022.1071731. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36618829 Free PMC article. Review.
-
Exploring Fatty Acid Mimetics as NR4A Ligands.J Med Chem. 2023 Nov 23;66(22):15362-15369. doi: 10.1021/acs.jmedchem.3c01467. Epub 2023 Nov 2. J Med Chem. 2023. PMID: 37918435 Free PMC article.
-
Flavone and Hydroxyflavones Are Ligands That Bind the Orphan Nuclear Receptor 4A1 (NR4A1).Int J Mol Sci. 2023 May 2;24(9):8152. doi: 10.3390/ijms24098152. Int J Mol Sci. 2023. PMID: 37175855 Free PMC article.
-
Toll-like receptors in sepsis-associated cytokine storm and their endogenous negative regulators as future immunomodulatory targets.Int Immunopharmacol. 2020 Dec;89(Pt B):107087. doi: 10.1016/j.intimp.2020.107087. Epub 2020 Oct 12. Int Immunopharmacol. 2020. PMID: 33075714 Free PMC article. Review.
-
The Osteogenesis Imperfecta Type V Mutant BRIL/IFITM5 Promotes Transcriptional Activation of MEF2, NFATc, and NR4A in Osteoblasts.Int J Mol Sci. 2022 Feb 15;23(4):2148. doi: 10.3390/ijms23042148. Int J Mol Sci. 2022. PMID: 35216266 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

