Cigarette Smoke Exposure and Inflammatory Signaling Increase the Expression of the SARS-CoV-2 Receptor ACE2 in the Respiratory Tract
- PMID: 32425701
- PMCID: PMC7229915
- DOI: 10.1016/j.devcel.2020.05.012
Cigarette Smoke Exposure and Inflammatory Signaling Increase the Expression of the SARS-CoV-2 Receptor ACE2 in the Respiratory Tract
Abstract
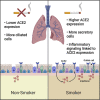
The factors mediating fatal SARS-CoV-2 infections are poorly understood. Here, we show that cigarette smoke causes a dose-dependent upregulation of angiotensin converting enzyme 2 (ACE2), the SARS-CoV-2 receptor, in rodent and human lungs. Using single-cell sequencing data, we demonstrate that ACE2 is expressed in a subset of secretory cells in the respiratory tract. Chronic smoke exposure triggers the expansion of this cell population and a concomitant increase in ACE2 expression. In contrast, quitting smoking decreases the abundance of these secretory cells and reduces ACE2 levels. Finally, we demonstrate that ACE2 expression is responsive to inflammatory signaling and can be upregulated by viral infections or interferon treatment. Taken together, these results may partially explain why smokers are particularly susceptible to severe SARS-CoV-2 infections. Furthermore, our work identifies ACE2 as an interferon-stimulated gene in lung cells, suggesting that SARS-CoV-2 infections could create positive feedback loops that increase ACE2 levels and facilitate viral dissemination.
Keywords: ACE2; COVID-19; Coronavirus; SARS-CoV-2; inflammation; interferon; lung development; smoking.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests J.C.S. is a co-founder of Meliora Therapeutics and is an employee of Google, Inc. This work was performed outside of her affiliation with Google and used no proprietary knowledge or materials from Google. J.M.S. has received consulting fees from Ono Pharmaceuticals, is a member of the Advisory Board of Tyra Biosciences, and is a co-founder of Meliora Therapeutics.
Figures




Similar articles
-
Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article.
-
SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues.Cell. 2020 May 28;181(5):1016-1035.e19. doi: 10.1016/j.cell.2020.04.035. Epub 2020 Apr 27. Cell. 2020. PMID: 32413319 Free PMC article.
-
Elevated FiO2 increases SARS-CoV-2 co-receptor expression in respiratory tract epithelium.Am J Physiol Lung Cell Mol Physiol. 2020 Oct 1;319(4):L670-L674. doi: 10.1152/ajplung.00345.2020. Epub 2020 Sep 2. Am J Physiol Lung Cell Mol Physiol. 2020. PMID: 32878480 Free PMC article.
-
Outcomes in Patients with COVID-19 Infection Taking ACEI/ARB.Curr Cardiol Rep. 2020 Apr 14;22(5):31. doi: 10.1007/s11886-020-01291-4. Curr Cardiol Rep. 2020. PMID: 32291526 Free PMC article. Review.
-
Two important controversial risk factors in SARS-CoV-2 infection: Obesity and smoking.Environ Toxicol Pharmacol. 2020 Aug;78:103411. doi: 10.1016/j.etap.2020.103411. Epub 2020 May 15. Environ Toxicol Pharmacol. 2020. PMID: 32422280 Free PMC article. Review.
Cited by
-
Aging, tobacco use and lung damages.Tunis Med. 2022 avril;100(4):295-302. Tunis Med. 2022. PMID: 36155900 Free PMC article.
-
Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity.bioRxiv [Preprint]. 2020 Aug 5:2020.08.05.237651. doi: 10.1101/2020.08.05.237651. bioRxiv. 2020. Update in: EBioMedicine. 2021 Mar;65:103255. doi: 10.1016/j.ebiom.2021.103255 PMID: 32793911 Free PMC article. Updated. Preprint.
-
Diseased lungs may hinder COVID-19 development: A possible reason for the low prevalence of COPD in COVID-19 patients.Med Hypotheses. 2021 Aug;153:110628. doi: 10.1016/j.mehy.2021.110628. Epub 2021 Jun 9. Med Hypotheses. 2021. PMID: 34139599 Free PMC article.
-
SARS-CoV-2 infection and smoking: What is the association? A brief review.Comput Struct Biotechnol J. 2021;19:1654-1660. doi: 10.1016/j.csbj.2021.03.023. Epub 2021 Mar 23. Comput Struct Biotechnol J. 2021. PMID: 33777332 Free PMC article. Review.
-
Sex Hormones and Hormone Therapy during COVID-19 Pandemic: Implications for Patients with Cancer.Cancers (Basel). 2020 Aug 18;12(8):2325. doi: 10.3390/cancers12082325. Cancers (Basel). 2020. PMID: 32824674 Free PMC article.
References
-
- Arnson Y., Shoenfeld Y., Amital H. Effects of tobacco smoke on immunity, inflammation and autoimmunity. J. Autoimmun. 2010;34:J258–J265. - PubMed
-
- Baumgartner K.B., Samet J.M., Stidley C.A., Colby T.V., Waldron J.A. Cigarette smoking: a risk factor for idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 1997;155:242–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

