Zoonotic origins of human coronaviruses
- PMID: 32226286
- PMCID: PMC7098031
- DOI: 10.7150/ijbs.45472
Zoonotic origins of human coronaviruses
Abstract
Mutation and adaptation have driven the co-evolution of coronaviruses (CoVs) and their hosts, including human beings, for thousands of years. Before 2003, two human CoVs (HCoVs) were known to cause mild illness, such as common cold. The outbreaks of severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) have flipped the coin to reveal how devastating and life-threatening an HCoV infection could be. The emergence of SARS-CoV-2 in central China at the end of 2019 has thrusted CoVs into the spotlight again and surprised us with its high transmissibility but reduced pathogenicity compared to its sister SARS-CoV. HCoV infection is a zoonosis and understanding the zoonotic origins of HCoVs would serve us well. Most HCoVs originated from bats where they are non-pathogenic. The intermediate reservoir hosts of some HCoVs are also known. Identifying the animal hosts has direct implications in the prevention of human diseases. Investigating CoV-host interactions in animals might also derive important insight on CoV pathogenesis in humans. In this review, we present an overview of the existing knowledge about the seven HCoVs, with a focus on the history of their discovery as well as their zoonotic origins and interspecies transmission. Importantly, we compare and contrast the different HCoVs from a perspective of virus evolution and genome recombination. The current CoV disease 2019 (COVID-19) epidemic is discussed in this context. In addition, the requirements for successful host switches and the implications of virus evolution on disease severity are also highlighted.
Keywords: COVID-19; MERS-CoV; SARS-CoV; SARS-CoV-2; animal reservoir; bats; coronavirus.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
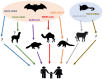
Similar articles
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
-
Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments.Vet Q. 2020 Dec;40(1):68-76. doi: 10.1080/01652176.2020.1727993. Vet Q. 2020. PMID: 32036774 Free PMC article. Review.
-
Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
-
Zoonotic and reverse zoonotic events of SARS-CoV-2 and their impact on global health.Emerg Microbes Infect. 2020 Dec;9(1):2222-2235. doi: 10.1080/22221751.2020.1827984. Emerg Microbes Infect. 2020. PMID: 32967592 Free PMC article. Review.
-
Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins.Nature. 2020 Jul;583(7815):282-285. doi: 10.1038/s41586-020-2169-0. Epub 2020 Mar 26. Nature. 2020. PMID: 32218527
Cited by
-
Addressing Biodisaster X Threats With Artificial Intelligence and 6G Technologies: Literature Review and Critical Insights.J Med Internet Res. 2021 May 25;23(5):e26109. doi: 10.2196/26109. J Med Internet Res. 2021. PMID: 33961583 Free PMC article. Review.
-
Machine learning methods accurately predict host specificity of coronaviruses based on spike sequences alone.Biochem Biophys Res Commun. 2020 Dec 10;533(3):553-558. doi: 10.1016/j.bbrc.2020.09.010. Epub 2020 Sep 18. Biochem Biophys Res Commun. 2020. PMID: 32981683 Free PMC article.
-
The diagnostic value of detecting sudden smell loss among asymptomatic COVID-19 patients in early stage: The possible early sign of COVID-19.Auris Nasus Larynx. 2020 Aug;47(4):565-573. doi: 10.1016/j.anl.2020.05.020. Epub 2020 Jun 9. Auris Nasus Larynx. 2020. PMID: 32553562 Free PMC article. Review.
-
[Virological aspects and diagnosis of SARS-CoV-2 coronavirus].Actual Pharm. 2020 Oct;59(599):18-23. doi: 10.1016/j.actpha.2020.08.005. Epub 2020 Aug 21. Actual Pharm. 2020. PMID: 32863557 Free PMC article. French.
-
COVID-19 and cardiovascular diseases.J Cardiol. 2020 Nov;76(5):453-458. doi: 10.1016/j.jjcc.2020.07.013. Epub 2020 Jul 22. J Cardiol. 2020. PMID: 32736906 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

