Transcriptomic and metabolomic analysis provides insights into anthocyanin and procyanidin accumulation in pear
- PMID: 32220242
- PMCID: PMC7099803
- DOI: 10.1186/s12870-020-02344-0
Transcriptomic and metabolomic analysis provides insights into anthocyanin and procyanidin accumulation in pear
Abstract
Background: Pear is one of the most important fruit crops worldwide. Anthocyanins and procyanidins (PAs) are important secondary metabolites that affect the appearance and nutritive quality of pear. However, few studies have focused on the molecular mechanism underlying anthocyanin and PA accumulation in pear.
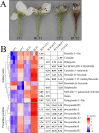
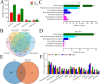
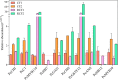
Results: We conducted metabolome and transcriptome analyses to identify candidate genes involved in anthocyanin and PA accumulation in young fruits of the pear cultivar 'Clapp Favorite' (CF) and its red mutation cultivar 'Red Clapp Favorite' (RCF). Gene-metabolite correlation analyses revealed a 'core set' of 20 genes that were strongly correlated with 10 anthocyanin and seven PA metabolites. Of these, PcGSTF12 was confirmed to be involved in anthocyanin and PA accumulation by complementation of the tt19-7 Arabidopsis mutant. Interestingly, PcGSTF12 was found to be responsible for the accumulation of procyanidin A3, but not petunidin 3, 5-diglucoside, opposite to the function of AtGSTs in Arabidopsis. Transformation with PcGSTF12 greatly promoted or repressed genes involved in anthocyanin and PA biosynthesis, regulation, and transport. Electrophoretic mobility shift and luciferase reporter assays confirmed positive regulation of PcGSTF12 by PcMYB114.
Conclusion: These findings identify a core set of genes for anthocyanin and PA accumulation in pear. Of these, PcGSTF12, was confirmed to be involved in anthocyanin and PA accumulation. Our results also identified an important anthocyanin and PA regulation node comprising two core genes, PcGSTF12 and PcMYB114. These results provide novel insights into anthocyanin and PA accumulation in pear and represent a valuable data set to guide future functional studies and pear breeding.
Keywords: Anthocyanin; Metabolome and transcriptome analyses; PcGSTF12; PcMYB114; Pear; Procyanidin.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Integrated metabolomic and transcriptomic analysis of the anthocyanin and proanthocyanidin regulatory networks in red walnut natural hybrid progeny leaves.PeerJ. 2022 Oct 20;10:e14262. doi: 10.7717/peerj.14262. eCollection 2022. PeerJ. 2022. PMID: 36285329 Free PMC article.
-
Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in 'Red Zaosu' pear fruits by interacting with MYB114.Plant Mol Biol. 2019 Jan;99(1-2):67-78. doi: 10.1007/s11103-018-0802-1. Epub 2018 Dec 11. Plant Mol Biol. 2019. PMID: 30539403
-
The blue light signal transduction pathway is involved in anthocyanin accumulation in 'Red Zaosu' pear.Planta. 2018 Jul;248(1):37-48. doi: 10.1007/s00425-018-2877-y. Epub 2018 Mar 15. Planta. 2018. PMID: 29546452
-
Regulatory Mechanisms of Anthocyanin Biosynthesis in Apple and Pear.Int J Mol Sci. 2021 Aug 6;22(16):8441. doi: 10.3390/ijms22168441. Int J Mol Sci. 2021. PMID: 34445149 Free PMC article. Review.
-
Biosynthetic Pathway of Proanthocyanidins in Major Cash Crops.Plants (Basel). 2021 Aug 28;10(9):1792. doi: 10.3390/plants10091792. Plants (Basel). 2021. PMID: 34579325 Free PMC article. Review.
Cited by
-
Feruloyl Glyceride Mitigates Tomato Postharvest Rot by Inhibiting Penicillium expansum Spore Germination and Enhancing Suberin Accumulation.Foods. 2024 Apr 10;13(8):1147. doi: 10.3390/foods13081147. Foods. 2024. PMID: 38672820 Free PMC article.
-
Tartary Buckwheat R2R3-MYB Gene FtMYB3 Negatively Regulates Anthocyanin and Proanthocyanin Biosynthesis.Int J Mol Sci. 2022 Mar 3;23(5):2775. doi: 10.3390/ijms23052775. Int J Mol Sci. 2022. PMID: 35269917 Free PMC article.
-
Molecular and Metabolic Insights into Anthocyanin Biosynthesis for Leaf Color Change in Chokecherry (Padus virginiana).Int J Mol Sci. 2021 Oct 2;22(19):10697. doi: 10.3390/ijms221910697. Int J Mol Sci. 2021. PMID: 34639038 Free PMC article.
-
Metabolomics Combined with Transcriptomics Analysis Revealed the Amino Acids, Phenolic Acids, and Flavonol Derivatives Biosynthesis Network in Developing Rosa roxburghii Fruit.Foods. 2022 Jun 1;11(11):1639. doi: 10.3390/foods11111639. Foods. 2022. PMID: 35681389 Free PMC article.
-
Functional R2R3-MYB transcription factor NsMYB1, regulating anthocyanin biosynthesis, was relative to the fruit color differentiation in Nitraria sibirica Pall.BMC Plant Biol. 2022 Apr 9;22(1):186. doi: 10.1186/s12870-022-03561-5. BMC Plant Biol. 2022. PMID: 35395726 Free PMC article.
References
-
- Ngo T, Zhao Y. Stabilization of anthocyanins on thermally processed red D’Anjou pears through complexation and polymerization. Food Sci Technol. 2009;42:1144–1152.
-
- Zhai R, Wang Z, Zhang S, Meng G, Song L, Wang Z, et al. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.) J Exp Bot. 2016;67:1275–1284. - PubMed
-
- Lesschaeve I, Noble AC. Polyphenols: factors influencing their sensory properties and their effects on food and beverage preferences. Am J Clin Nutr. 2005;81:330–335. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

