Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine
- PMID: 32203189
- PMCID: PMC7091888
- DOI: 10.1038/s41423-020-0400-4
Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine
Abstract
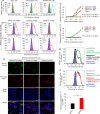
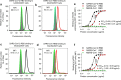
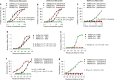
The outbreak of Coronavirus Disease 2019 (COVID-19) has posed a serious threat to global public health, calling for the development of safe and effective prophylactics and therapeutics against infection of its causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as 2019 novel coronavirus (2019-nCoV). The CoV spike (S) protein plays the most important roles in viral attachment, fusion and entry, and serves as a target for development of antibodies, entry inhibitors and vaccines. Here, we identified the receptor-binding domain (RBD) in SARS-CoV-2 S protein and found that the RBD protein bound strongly to human and bat angiotensin-converting enzyme 2 (ACE2) receptors. SARS-CoV-2 RBD exhibited significantly higher binding affinity to ACE2 receptor than SARS-CoV RBD and could block the binding and, hence, attachment of SARS-CoV-2 RBD and SARS-CoV RBD to ACE2-expressing cells, thus inhibiting their infection to host cells. SARS-CoV RBD-specific antibodies could cross-react with SARS-CoV-2 RBD protein, and SARS-CoV RBD-induced antisera could cross-neutralize SARS-CoV-2, suggesting the potential to develop SARS-CoV RBD-based vaccines for prevention of SARS-CoV-2 and SARS-CoV infection.
Keywords: 2019 novel coronavirus; SARS-CoV-2; cross-neutralization; receptor-binding domain; spike protein; viral inhibitor.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts.J Virol. 2020 Jul 16;94(15):e00831-20. doi: 10.1128/JVI.00831-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32404529 Free PMC article.
-
Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2.Antiviral Res. 2020 Jul;179:104820. doi: 10.1016/j.antiviral.2020.104820. Epub 2020 May 13. Antiviral Res. 2020. PMID: 32405117 Free PMC article.
-
Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies.Cell Mol Immunol. 2020 Jun;17(6):621-630. doi: 10.1038/s41423-020-0458-z. Epub 2020 May 15. Cell Mol Immunol. 2020. PMID: 32415260 Free PMC article.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
-
The SARS-CoV-2 Spike Glycoprotein as a Drug and Vaccine Target: Structural Insights into Its Complexes with ACE2 and Antibodies.Cells. 2020 Oct 22;9(11):2343. doi: 10.3390/cells9112343. Cells. 2020. PMID: 33105869 Free PMC article. Review.
Cited by
-
NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies.EMBO Rep. 2021 May 5;22(5):e52325. doi: 10.15252/embr.202052325. Epub 2021 Apr 27. EMBO Rep. 2021. PMID: 33904225 Free PMC article.
-
Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine-Specific Cancer Targeting with Undetectable Toxicity.Chem Rev. 2021 Jul 14;121(13):7398-7467. doi: 10.1021/acs.chemrev.1c00009. Epub 2021 May 26. Chem Rev. 2021. PMID: 34038115 Free PMC article. Review.
-
African swine fever virus modulates the endoplasmic reticulum stress-ATF6-calcium axis to facilitate viral replication.Emerg Microbes Infect. 2024 Dec;13(1):2399945. doi: 10.1080/22221751.2024.2399945. Epub 2024 Sep 27. Emerg Microbes Infect. 2024. PMID: 39230190 Free PMC article.
-
Features of the Humoral Response to Infection, Vaccination, and Revaccination during COVID-19.Bull Exp Biol Med. 2022 Oct;173(6):734-739. doi: 10.1007/s10517-022-05620-1. Epub 2022 Nov 2. Bull Exp Biol Med. 2022. PMID: 36322302 Free PMC article.
-
Strategies for Immunomonitoring after Vaccination and during Infection.Vaccines (Basel). 2021 Apr 9;9(4):365. doi: 10.3390/vaccines9040365. Vaccines (Basel). 2021. PMID: 33918841 Free PMC article. Review.
References
-
- World Health Organization. Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technica... (2020).
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI137472/AI/NIAID NIH HHS/United States
- R01 AI139092/AI/NIAID NIH HHS/United States
- R01AI139092/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
- R01AI137472/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

