Drug targets for corona virus: A systematic review
- PMID: 32201449
- PMCID: PMC7074424
- DOI: 10.4103/ijp.IJP_115_20
Drug targets for corona virus: A systematic review
Abstract
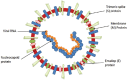
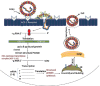
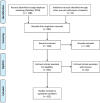
The 2019-novel coronavirus (nCoV) is a major source of disaster in the 21th century. However, the lack of specific drugs to prevent/treat an attack is a major need at this current point of time. In this regard, we conducted a systematic review to identify major druggable targets in coronavirus (CoV). We searched PubMed and RCSB database with keywords HCoV, NCoV, corona virus, SERS-CoV, MERS-CoV, 2019-nCoV, crystal structure, X-ray crystallography structure, NMR structure, target, and drug target till Feb 3, 2020. The search identified seven major targets (spike protein, envelop protein, membrane protein, protease, nucleocapsid protein, hemagglutinin esterase, and helicase) for which drug design can be considered. There are other 16 nonstructural proteins (NSPs), which can also be considered from the drug design perspective. The major structural proteins and NSPs may serve an important role from drug design perspectives. However, the occurrence of frequent recombination events is a major deterrent factor toward the development of CoV-specific vaccines/drugs.
Keywords: Coronavirus; Middle East respiratory syndrome; drug targets; severe acute respiratory syndrome.
Copyright: © 2020 Indian Journal of Pharmacology.
Conflict of interest statement
There are no conflicts of interest.
Figures
Similar articles
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
-
SARS, MERS and SARS-CoV-2 (COVID-19) treatment: a patent review.Expert Opin Ther Pat. 2020 Aug;30(8):567-579. doi: 10.1080/13543776.2020.1772231. Epub 2020 Jun 7. Expert Opin Ther Pat. 2020. PMID: 32429703 Review.
-
Update on the target structures of SARS-CoV-2: A systematic review.Indian J Pharmacol. 2020 Mar-Apr;52(2):142-149. doi: 10.4103/ijp.IJP_338_20. Epub 2020 Jun 3. Indian J Pharmacol. 2020. PMID: 32565603 Free PMC article.
-
Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19).Asian Pac J Allergy Immunol. 2020 Mar;38(1):10-18. doi: 10.12932/AP-200220-0773. Asian Pac J Allergy Immunol. 2020. PMID: 32134278 Review.
-
Corona virus versus existence of human on the earth: A computational and biophysical approach.Int J Biol Macromol. 2020 Oct 15;161:271-281. doi: 10.1016/j.ijbiomac.2020.06.007. Epub 2020 Jun 5. Int J Biol Macromol. 2020. PMID: 32512089 Free PMC article. Review.
Cited by
-
Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: a pharmacoinformatics study.J Biomol Struct Dyn. 2021 Aug;39(13):4686-4700. doi: 10.1080/07391102.2020.1779132. Epub 2020 Jun 18. J Biomol Struct Dyn. 2021. PMID: 32552462 Free PMC article.
-
Multi-targeted molecular docking, pharmacokinetics, and drug-likeness evaluation of coumarin based compounds targeting proteins involved in development of COVID-19.Saudi J Biol Sci. 2022 Dec;29(12):103458. doi: 10.1016/j.sjbs.2022.103458. Epub 2022 Sep 27. Saudi J Biol Sci. 2022. PMID: 36187455 Free PMC article.
-
Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of ACEIs Against SARS-CoV-2 Targeting the hACE2 Receptor.Front Chem. 2021 May 4;9:661230. doi: 10.3389/fchem.2021.661230. eCollection 2021. Front Chem. 2021. PMID: 34017819 Free PMC article.
-
Antiviral potential of nanoparticles for the treatment of Coronavirus infections.J Trace Elem Med Biol. 2022 Jul;72:126977. doi: 10.1016/j.jtemb.2022.126977. Epub 2022 Mar 26. J Trace Elem Med Biol. 2022. PMID: 35397331 Free PMC article. Review.
-
Computational and in vitro experimental analyses of the anti-COVID-19 potential of Mortaparib and MortaparibPlus.Biosci Rep. 2021 Oct 29;41(10):BSR20212156. doi: 10.1042/BSR20212156. Biosci Rep. 2021. PMID: 34647577 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

