Erythromyeloid progenitors give rise to a population of osteoclasts that contribute to bone homeostasis and repair
- PMID: 31907410
- PMCID: PMC6953622
- DOI: 10.1038/s41556-019-0437-8
Erythromyeloid progenitors give rise to a population of osteoclasts that contribute to bone homeostasis and repair
Abstract
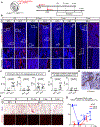
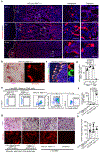
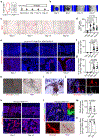
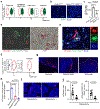
Osteoclasts are multinucleated cells of the monocyte/macrophage lineage that degrade bone. Here, we used lineage tracing studies-labelling cells expressing Cx3cr1, Csf1r or Flt3-to identify the precursors of osteoclasts in mice. We identified an erythromyeloid progenitor (EMP)-derived osteoclast precursor population. Yolk-sac macrophages of EMP origin produced neonatal osteoclasts that can create a space for postnatal bone marrow haematopoiesis. Furthermore, EMPs gave rise to long-lasting osteoclast precursors that contributed to postnatal bone remodelling in both physiological and pathological settings. Our single-cell RNA-sequencing data showed that EMP-derived osteoclast precursors arose independently of the haematopoietic stem cell (HSC) lineage and the data from fate tracking of EMP and HSC lineages indicated the possibility of cell-cell fusion between these two lineages. Cx3cr1+ yolk-sac macrophage descendants resided in the adult spleen, and parabiosis experiments showed that these cells migrated through the bloodstream to the remodelled bone after injury.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures






Similar articles
-
Monocyte/Macrophage Lineage Cells From Fetal Erythromyeloid Progenitors Orchestrate Bone Remodeling and Repair.Front Cell Dev Biol. 2021 Feb 4;9:622035. doi: 10.3389/fcell.2021.622035. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33614650 Free PMC article. Review.
-
Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors.Nature. 2015 Feb 26;518(7540):547-51. doi: 10.1038/nature13989. Epub 2014 Dec 3. Nature. 2015. PMID: 25470051 Free PMC article.
-
Identification Of Erythromyeloid Progenitors And Their Progeny In The Mouse Embryo By Flow Cytometry.J Vis Exp. 2017 Jul 17;(125):55305. doi: 10.3791/55305. J Vis Exp. 2017. PMID: 28745620 Free PMC article.
-
Megakaryocyte production is sustained by direct differentiation from erythromyeloid progenitors in the yolk sac until midgestation.Immunity. 2021 Jul 13;54(7):1433-1446.e5. doi: 10.1016/j.immuni.2021.04.026. Epub 2021 May 31. Immunity. 2021. PMID: 34062116 Free PMC article.
-
Fetal monocytes and the origins of tissue-resident macrophages.Cell Immunol. 2018 Aug;330:5-15. doi: 10.1016/j.cellimm.2018.01.001. Epub 2018 Jan 12. Cell Immunol. 2018. PMID: 29475558 Review.
Cited by
-
The M-CSF receptor in osteoclasts and beyond.Exp Mol Med. 2020 Aug;52(8):1239-1254. doi: 10.1038/s12276-020-0484-z. Epub 2020 Aug 17. Exp Mol Med. 2020. PMID: 32801364 Free PMC article. Review.
-
Asiatic Acid Attenuates Osteoporotic Bone Loss in Ovariectomized Mice Through Inhibiting NF-kappaB/MAPK/ Protein Kinase B Signaling Pathway.Front Pharmacol. 2022 Feb 8;13:829741. doi: 10.3389/fphar.2022.829741. eCollection 2022. Front Pharmacol. 2022. PMID: 35211021 Free PMC article.
-
NanoZnO-modified titanium implants for enhanced anti-bacterial activity, osteogenesis and corrosion resistance.J Nanobiotechnology. 2021 Oct 30;19(1):353. doi: 10.1186/s12951-021-01099-6. J Nanobiotechnology. 2021. PMID: 34717648 Free PMC article. Review.
-
IFT20 and WWTR1 govern bone homeostasis via synchronously regulating the expression and stability of TβRII in osteoblast lineage cells.Res Sq [Preprint]. 2024 Mar 19:rs.3.rs-4009802. doi: 10.21203/rs.3.rs-4009802/v1. Res Sq. 2024. PMID: 38562782 Free PMC article. Preprint.
-
Two macrophages, osteoclasts and microglia: from development to pleiotropy.Bone Res. 2021 Feb 10;9(1):11. doi: 10.1038/s41413-020-00134-w. Bone Res. 2021. PMID: 33568650 Free PMC article. Review.
References
-
- Frost HM, Vilanueva AR, Jett S & Eyring E Tetracycline-based analysis of bone remodelling in osteopetrosis. Clinical orthopaedics and related research 65, 203–217 (1969). - PubMed
-
- Yue R, Zhou Bo O. Shimada, Issei S, Zhao Z & Morrison Sean J. Leptin Receptor Promotes Adipogenesis and Reduces Osteogenesis by Regulating Mesenchymal Stromal Cells in Adult Bone Marrow. Cell Stem Cell 18, 782–796 (2016). - PubMed
-
- Udagawa N et al. Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proceedings of the National Academy of Sciences of the United States of America 87, 7260–7264 (1990). - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

