Investigation of a dilated cardiomyopathy-associated variant in BAG3 using genome-edited iPSC-derived cardiomyocytes
- PMID: 31723063
- PMCID: PMC6948852
- DOI: 10.1172/jci.insight.128799
Investigation of a dilated cardiomyopathy-associated variant in BAG3 using genome-edited iPSC-derived cardiomyocytes
Abstract
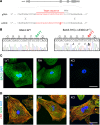
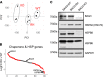
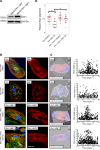
Mutations in B cell lymphoma 2-associated athanogene 3 (BAG3) are recurrently associated with dilated cardiomyopathy (DCM) and muscular dystrophy. Using isogenic genome-edited human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs), we examined how a DCM-causing BAG3 mutation (R477H), as well as complete loss of BAG3 (KO), impacts myofibrillar organization and chaperone networks. Although unchanged at baseline, fiber length and alignment declined markedly in R477H and KO iPSC-CMs following proteasome inhibition. RNA sequencing revealed extensive baseline changes in chaperone- and stress response protein-encoding genes, and protein levels of key BAG3 binding partners were perturbed. Molecular dynamics simulations of the BAG3-HSC70 complex predicted a partial disengagement by the R477H mutation. In line with this, BAG3-R477H bound less HSC70 than BAG3-WT in coimmunoprecipitation assays. Finally, myofibrillar disarray triggered by proteasome inhibition in R477H cells was mitigated by overexpression of the stress response protein heat shock factor 1 (HSF1). These studies reveal the importance of BAG3 in coordinating protein quality control subsystem usage within the cardiomyocyte and suggest that augmenting HSF1 activity might be beneficial as a means to mitigate proteostatic stress in the context of BAG3-associated DCM.
Keywords: Cardiology; Genetics; Heart failure; iPS cells.
Conflict of interest statement
Figures


Similar articles
-
Advances in the role and mechanism of BAG3 in dilated cardiomyopathy.Heart Fail Rev. 2021 Jan;26(1):183-194. doi: 10.1007/s10741-019-09899-7. Heart Fail Rev. 2021. PMID: 31808029 Review.
-
Generation of two human iPSC lines with Exon 3 mutations in BCL2-Associated Athanogene 3 (BAG3) from dilated cardiomyopathy patients.Stem Cell Res. 2023 Mar;67:103019. doi: 10.1016/j.scr.2023.103019. Epub 2023 Jan 5. Stem Cell Res. 2023. PMID: 36642055 Free PMC article.
-
BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress.Circ Res. 2010 Nov 12;107(10):1220-31. doi: 10.1161/CIRCRESAHA.110.225649. Epub 2010 Sep 30. Circ Res. 2010. PMID: 20884878 Free PMC article.
-
Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes.Hum Mutat. 2011 Dec;32(12):1481-91. doi: 10.1002/humu.21603. Epub 2011 Sep 29. Hum Mutat. 2011. PMID: 21898660
-
The role of BAG3 in dilated cardiomyopathy and its association with Charcot-Marie-Tooth disease type 2.Acta Myol. 2022 Jun 30;41(2):59-75. doi: 10.36185/2532-1900-071. eCollection 2022 Jun. Acta Myol. 2022. PMID: 35832504 Free PMC article. Review.
Cited by
-
Machine learning and bioinformatics to identify 8 autophagy-related biomarkers and construct gene regulatory networks in dilated cardiomyopathy.Sci Rep. 2022 Sep 2;12(1):15030. doi: 10.1038/s41598-022-19027-5. Sci Rep. 2022. PMID: 36056063 Free PMC article.
-
Dysregulated Autophagy and Sarcomere Dysfunction in Patients With Heart Failure With Co-Occurrence of P63A and P380S BAG3 Variants.J Am Heart Assoc. 2023 Dec 19;12(24):e029938. doi: 10.1161/JAHA.123.029938. Epub 2023 Dec 18. J Am Heart Assoc. 2023. PMID: 38108245 Free PMC article.
-
Dynamic effects of genetic variation on gene expression revealed following hypoxic stress in cardiomyocytes.Elife. 2021 Feb 8;10:e57345. doi: 10.7554/eLife.57345. Elife. 2021. PMID: 33554857 Free PMC article.
-
Genome Editing for the Understanding and Treatment of Inherited Cardiomyopathies.Int J Mol Sci. 2020 Jan 22;21(3):733. doi: 10.3390/ijms21030733. Int J Mol Sci. 2020. PMID: 31979133 Free PMC article. Review.
-
Come Together: Protein Assemblies, Aggregates and the Sarcostat at the Heart of Cardiac Myocyte Homeostasis.Front Physiol. 2020 Jun 4;11:586. doi: 10.3389/fphys.2020.00586. eCollection 2020. Front Physiol. 2020. PMID: 32581848 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

